Prarabdh Shukla
Silencing Empowerment, Allowing Bigotry: Auditing the Moderation of Hate Speech on Twitch
Jun 09, 2025Abstract:To meet the demands of content moderation, online platforms have resorted to automated systems. Newer forms of real-time engagement($\textit{e.g.}$, users commenting on live streams) on platforms like Twitch exert additional pressures on the latency expected of such moderation systems. Despite their prevalence, relatively little is known about the effectiveness of these systems. In this paper, we conduct an audit of Twitch's automated moderation tool ($\texttt{AutoMod}$) to investigate its effectiveness in flagging hateful content. For our audit, we create streaming accounts to act as siloed test beds, and interface with the live chat using Twitch's APIs to send over $107,000$ comments collated from $4$ datasets. We measure $\texttt{AutoMod}$'s accuracy in flagging blatantly hateful content containing misogyny, racism, ableism and homophobia. Our experiments reveal that a large fraction of hateful messages, up to $94\%$ on some datasets, $\textit{bypass moderation}$. Contextual addition of slurs to these messages results in $100\%$ removal, revealing $\texttt{AutoMod}$'s reliance on slurs as a moderation signal. We also find that contrary to Twitch's community guidelines, $\texttt{AutoMod}$ blocks up to $89.5\%$ of benign examples that use sensitive words in pedagogical or empowering contexts. Overall, our audit points to large gaps in $\texttt{AutoMod}$'s capabilities and underscores the importance for such systems to understand context effectively.
DiffRed: Dimensionality Reduction guided by stable rank
Mar 09, 2024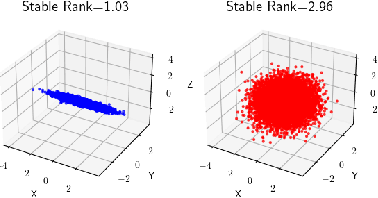
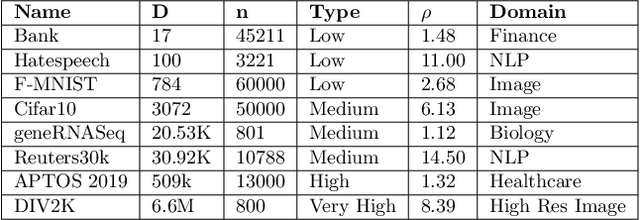
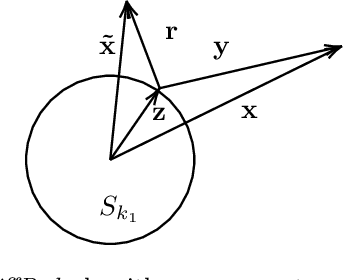
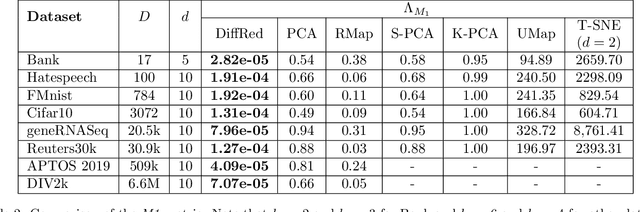
Abstract:In this work, we propose a novel dimensionality reduction technique, DiffRed, which first projects the data matrix, A, along first $k_1$ principal components and the residual matrix $A^{*}$ (left after subtracting its $k_1$-rank approximation) along $k_2$ Gaussian random vectors. We evaluate M1, the distortion of mean-squared pair-wise distance, and Stress, the normalized value of RMS of distortion of the pairwise distances. We rigorously prove that DiffRed achieves a general upper bound of $O\left(\sqrt{\frac{1-p}{k_2}}\right)$ on Stress and $O\left(\frac{(1-p)}{\sqrt{k_2*\rho(A^{*})}}\right)$ on M1 where $p$ is the fraction of variance explained by the first $k_1$ principal components and $\rho(A^{*})$ is the stable rank of $A^{*}$. These bounds are tighter than the currently known results for Random maps. Our extensive experiments on a variety of real-world datasets demonstrate that DiffRed achieves near zero M1 and much lower values of Stress as compared to the well-known dimensionality reduction techniques. In particular, DiffRed can map a 6 million dimensional dataset to 10 dimensions with 54% lower Stress than PCA.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge