Perry Siehien
An Evaluation of Real-time Adaptive Sampling Change Point Detection Algorithm using KCUSUM
Feb 15, 2024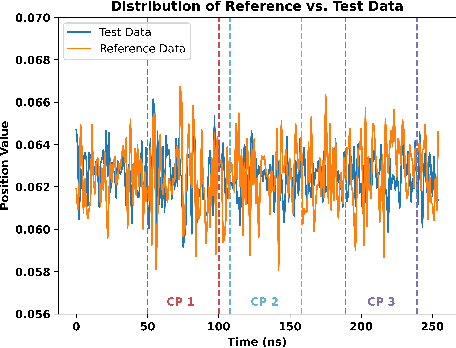

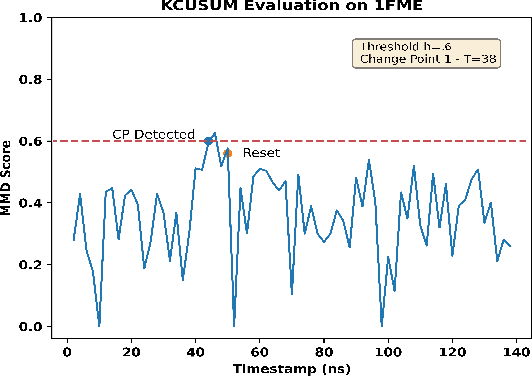
Abstract:Detecting abrupt changes in real-time data streams from scientific simulations presents a challenging task, demanding the deployment of accurate and efficient algorithms. Identifying change points in live data stream involves continuous scrutiny of incoming observations for deviations in their statistical characteristics, particularly in high-volume data scenarios. Maintaining a balance between sudden change detection and minimizing false alarms is vital. Many existing algorithms for this purpose rely on known probability distributions, limiting their feasibility. In this study, we introduce the Kernel-based Cumulative Sum (KCUSUM) algorithm, a non-parametric extension of the traditional Cumulative Sum (CUSUM) method, which has gained prominence for its efficacy in online change point detection under less restrictive conditions. KCUSUM splits itself by comparing incoming samples directly with reference samples and computes a statistic grounded in the Maximum Mean Discrepancy (MMD) non-parametric framework. This approach extends KCUSUM's pertinence to scenarios where only reference samples are available, such as atomic trajectories of proteins in vacuum, facilitating the detection of deviations from the reference sample without prior knowledge of the data's underlying distribution. Furthermore, by harnessing MMD's inherent random-walk structure, we can theoretically analyze KCUSUM's performance across various use cases, including metrics like expected delay and mean runtime to false alarms. Finally, we discuss real-world use cases from scientific simulations such as NWChem CODAR and protein folding data, demonstrating KCUSUM's practical effectiveness in online change point detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge