Partha Mitra
Overfitting or perfect fitting? Risk bounds for classification and regression rules that interpolate
Oct 26, 2018
Abstract:Many modern machine learning models are trained to achieve zero or near-zero training error in order to obtain near-optimal (but non-zero) test error. This phenomenon of strong generalization performance for "overfitted" / interpolated classifiers appears to be ubiquitous in high-dimensional data, having been observed in deep networks, kernel machines, boosting and random forests. Their performance is consistently robust even when the data contain large amounts of label noise. Very little theory is available to explain these observations. The vast majority of theoretical analyses of generalization allows for interpolation only when there is little or no label noise. This paper takes a step toward a theoretical foundation for interpolated classifiers by analyzing local interpolating schemes, including geometric simplicial interpolation algorithm and singularly weighted $k$-nearest neighbor schemes. Consistency or near-consistency is proved for these schemes in classification and regression problems. Moreover, the nearest neighbor schemes exhibit optimal rates under some standard statistical assumptions. Finally, this paper suggests a way to explain the phenomenon of adversarial examples, which are seemingly ubiquitous in modern machine learning, and also discusses some connections to kernel machines and random forests in the interpolated regime.
The Active Atlas: Combining 3D Anatomical Models with Texture Detectors
Jan 15, 2018


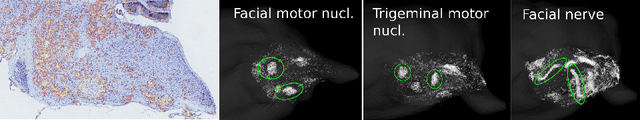
Abstract:While modern imaging technologies such as fMRI have opened exciting new possibilities for studying the brain in vivo, histological sections remain the best way to study the anatomy of the brain at the level of single neurons. The histological atlas changed little since 1909 and localizing brain regions is a still a labor intensive process performed only by experienced neuro-anatomists. Existing digital atlases such as the Allen Brain atlas are limited to low resolution images which cannot identify the detailed structure of the neurons. We have developed a digital atlas methodology that combines information about the 3D organization of the brain and the detailed texture of neurons in different structures. Using the methodology we developed an atlas for the mouse brainstem and mid-brain, two regions for which there are currently no good atlases. Our atlas is "active" in that it can be used to automatically align a histological stack to the atlas, thus reducing the work of the neuroanatomist.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge