Panos Antsaklis
Encoding Multi-Resolution Brain Networks Using Unsupervised Deep Learning
Aug 13, 2017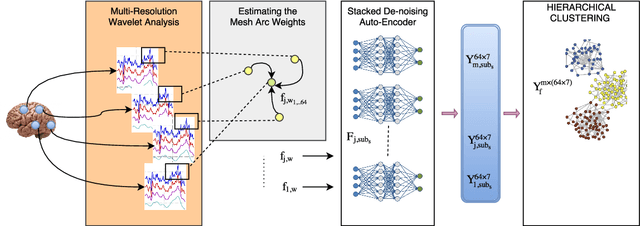
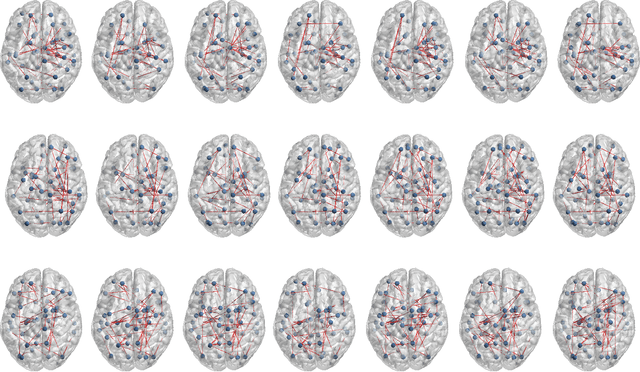
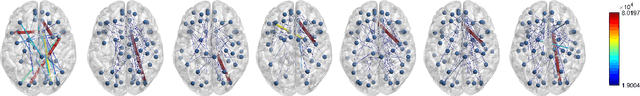

Abstract:The main goal of this study is to extract a set of brain networks in multiple time-resolutions to analyze the connectivity patterns among the anatomic regions for a given cognitive task. We suggest a deep architecture which learns the natural groupings of the connectivity patterns of human brain in multiple time-resolutions. The suggested architecture is tested on task data set of Human Connectome Project (HCP) where we extract multi-resolution networks, each of which corresponds to a cognitive task. At the first level of this architecture, we decompose the fMRI signal into multiple sub-bands using wavelet decompositions. At the second level, for each sub-band, we estimate a brain network extracted from short time windows of the fMRI signal. At the third level, we feed the adjacency matrices of each mesh network at each time-resolution into an unsupervised deep learning algorithm, namely, a Stacked De- noising Auto-Encoder (SDAE). The outputs of the SDAE provide a compact connectivity representation for each time window at each sub-band of the fMRI signal. We concatenate the learned representations of all sub-bands at each window and cluster them by a hierarchical algorithm to find the natural groupings among the windows. We observe that each cluster represents a cognitive task with a performance of 93% Rand Index and 71% Adjusted Rand Index. We visualize the mean values and the precisions of the networks at each component of the cluster mixture. The mean brain networks at cluster centers show the variations among cognitive tasks and the precision of each cluster shows the within cluster variability of networks, across the subjects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge