Panagiotis Kosmas
Non-Invasive Glucose Prediction System Enhanced by Mixed Linear Models and Meta-Forests for Domain Generalization
Sep 11, 2024Abstract:In this study, we present a non-invasive glucose prediction system that integrates Near-Infrared (NIR) spectroscopy and millimeter-wave (mm-wave) sensing. We employ a Mixed Linear Model (MixedLM) to analyze the association between mm-wave frequency S_21 parameters and blood glucose levels within a heterogeneous dataset. The MixedLM method considers inter-subject variability and integrates multiple predictors, offering a more comprehensive analysis than traditional correlation analysis. Additionally, we incorporate a Domain Generalization (DG) model, Meta-forests, to effectively handle domain variance in the dataset, enhancing the model's adaptability to individual differences. Our results demonstrate promising accuracy in glucose prediction for unseen subjects, with a mean absolute error (MAE) of 17.47 mg/dL, a root mean square error (RMSE) of 31.83 mg/dL, and a mean absolute percentage error (MAPE) of 10.88%, highlighting its potential for clinical application. This study marks a significant step towards developing accurate, personalized, and non-invasive glucose monitoring systems, contributing to improved diabetes management.
Exploring Biomarker Relationships in Both Type 1 and Type 2 Diabetes Mellitus Through a Bayesian Network Analysis Approach
Jun 24, 2024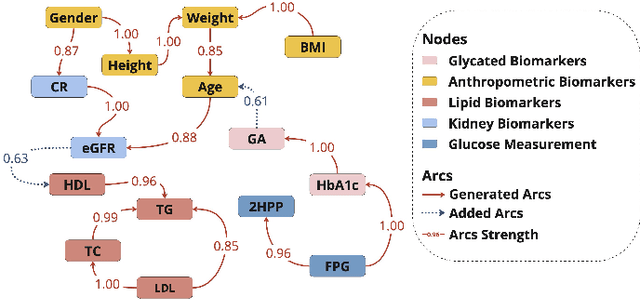
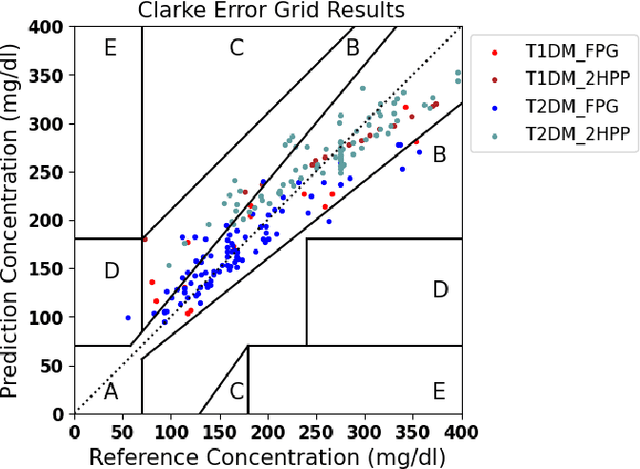
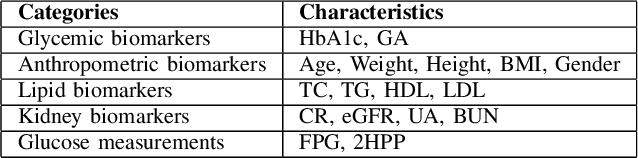
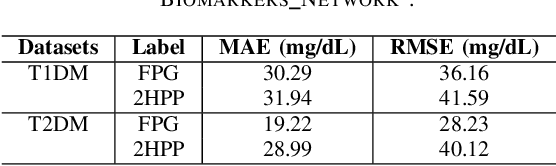
Abstract:Understanding the complex relationships of biomarkers in diabetes is pivotal for advancing treatment strategies, a pressing need in diabetes research. This study applies Bayesian network structure learning to analyze the Shanghai Type 1 and Type 2 diabetes mellitus datasets, revealing complex relationships among key diabetes-related biomarkers. The constructed Bayesian network presented notable predictive accuracy, particularly for Type 2 diabetes mellitus, with root mean squared error (RMSE) of 18.23 mg/dL, as validated through leave-one-domain experiments and Clarke error grid analysis. This study not only elucidates the intricate dynamics of diabetes through a deeper understanding of biomarker interplay but also underscores the significant potential of integrating data-driven and knowledge-driven methodologies in the realm of personalized diabetes management. Such an approach paves the way for more custom and effective treatment strategies, marking a notable advancement in the field.
Meta-forests: Domain generalization on random forests with meta-learning
Jan 09, 2024Abstract:Domain generalization is a popular machine learning technique that enables models to perform well on the unseen target domain, by learning from multiple source domains. Domain generalization is useful in cases where data is limited, difficult, or expensive to collect, such as in object recognition and biomedicine. In this paper, we propose a novel domain generalization algorithm called "meta-forests", which builds upon the basic random forests model by incorporating the meta-learning strategy and maximum mean discrepancy measure. The aim of meta-forests is to enhance the generalization ability of classifiers by reducing the correlation among trees and increasing their strength. More specifically, meta-forests conducts meta-learning optimization during each meta-task, while also utilizing the maximum mean discrepancy as a regularization term to penalize poor generalization performance in the meta-test process. To evaluate the effectiveness of our algorithm, we test it on two publicly object recognition datasets and a glucose monitoring dataset that we have used in a previous study. Our results show that meta-forests outperforms state-of-the-art approaches in terms of generalization performance on both object recognition and glucose monitoring datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge