Panagiotis Anagnostou
Detection of Fake Generated Scientific Abstracts
Apr 12, 2023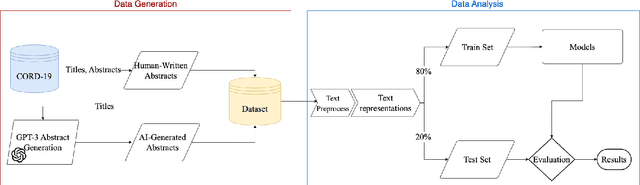
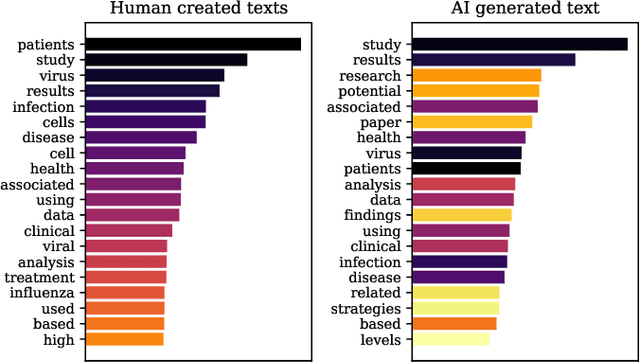
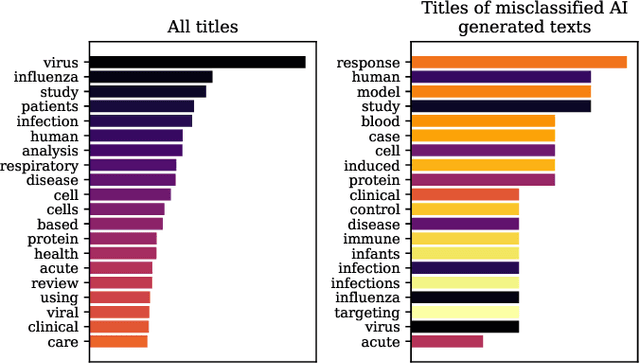
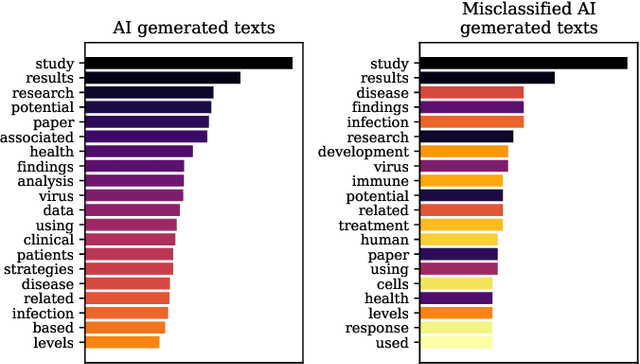
Abstract:The widespread adoption of Large Language Models and publicly available ChatGPT has marked a significant turning point in the integration of Artificial Intelligence into people's everyday lives. The academic community has taken notice of these technological advancements and has expressed concerns regarding the difficulty of discriminating between what is real and what is artificially generated. Thus, researchers have been working on developing effective systems to identify machine-generated text. In this study, we utilize the GPT-3 model to generate scientific paper abstracts through Artificial Intelligence and explore various text representation methods when combined with Machine Learning models with the aim of identifying machine-written text. We analyze the models' performance and address several research questions that rise during the analysis of the results. By conducting this research, we shed light on the capabilities and limitations of Artificial Intelligence generated text.
HiPart: Hierarchical Divisive Clustering Toolbox
Sep 18, 2022
Abstract:This paper presents the HiPart package, an open-source native python library that provides efficient and interpret-able implementations of divisive hierarchical clustering algorithms. HiPart supports interactive visualizations for the manipulation of the execution steps allowing the direct intervention of the clustering outcome. This package is highly suited for Big Data applications as the focus has been given to the computational efficiency of the implemented clustering methodologies. The dependencies used are either Python build-in packages or highly maintained stable external packages. The software is provided under the MIT license. The package's source code and documentation can be found at https://github.com/panagiotisanagnostou/HiPart.
Approximate kNN Classification for Biomedical Data
Dec 03, 2020
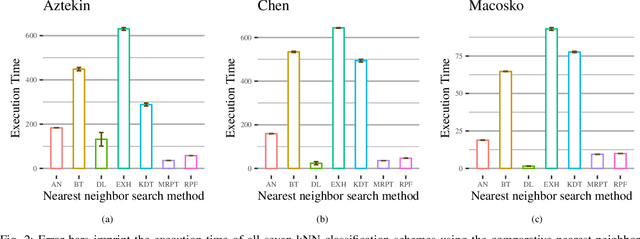
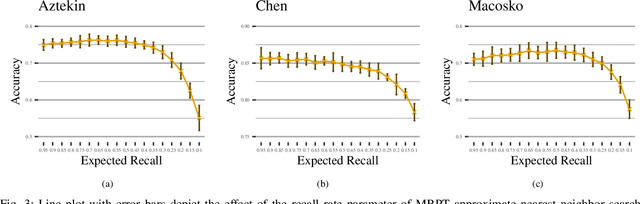
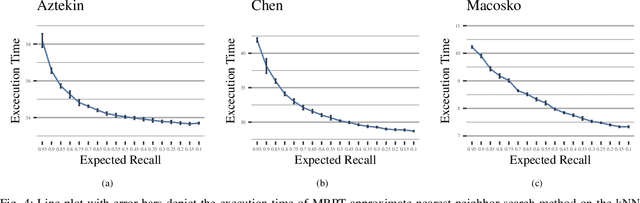
Abstract:We are in the era where the Big Data analytics has changed the way of interpreting the various biomedical phenomena, and as the generated data increase, the need for new machine learning methods to handle this evolution grows. An indicative example is the single-cell RNA-seq (scRNA-seq), an emerging DNA sequencing technology with promising capabilities but significant computational challenges due to the large-scaled generated data. Regarding the classification process for scRNA-seq data, an appropriate method is the k Nearest Neighbor (kNN) classifier since it is usually utilized for large-scale prediction tasks due to its simplicity, minimal parameterization, and model-free nature. However, the ultra-high dimensionality that characterizes scRNA-seq impose a computational bottleneck, while prediction power can be affected by the "Curse of Dimensionality". In this work, we proposed the utilization of approximate nearest neighbor search algorithms for the task of kNN classification in scRNA-seq data focusing on a particular methodology tailored for high dimensional data. We argue that even relaxed approximate solutions will not affect the prediction performance significantly. The experimental results confirm the original assumption by offering the potential for broader applicability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge