Aristidis G. Vrahatis
Graph Neural Network approaches for single-cell data: A recent overview
Oct 14, 2023


Abstract:Graph Neural Networks (GNN) are reshaping our understanding of biomedicine and diseases by revealing the deep connections among genes and cells. As both algorithmic and biomedical technologies have advanced significantly, we're entering a transformative phase of personalized medicine. While pioneering tools like Graph Attention Networks (GAT) and Graph Convolutional Neural Networks (Graph CNN) are advancing graph-based learning, the rise of single-cell sequencing techniques is reshaping our insights on cellular diversity and function. Numerous studies have combined GNNs with single-cell data, showing promising results. In this work, we highlight the GNN methodologies tailored for single-cell data over the recent years. We outline the diverse range of graph deep learning architectures that center on GAT methodologies. Furthermore, we underscore the several objectives of GNN strategies in single-cell data contexts, ranging from cell-type annotation, data integration and imputation, gene regulatory network reconstruction, clustering and many others. This review anticipates a future where GNNs become central to single-cell analysis efforts, particularly as vast omics datasets are continuously generated and the interconnectedness of cells and genes enhances our depth of knowledge in biomedicine.
Approximate kNN Classification for Biomedical Data
Dec 03, 2020
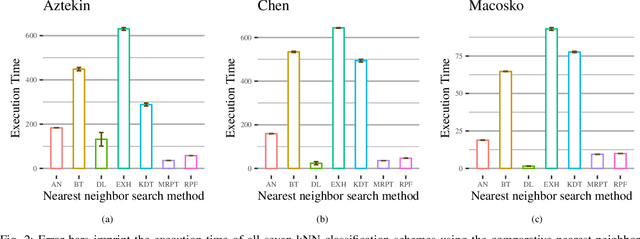
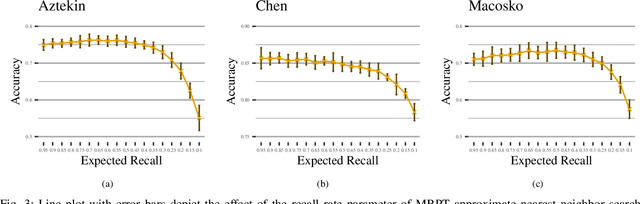
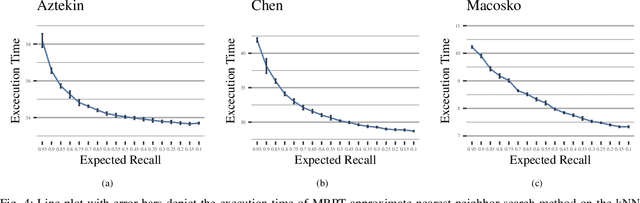
Abstract:We are in the era where the Big Data analytics has changed the way of interpreting the various biomedical phenomena, and as the generated data increase, the need for new machine learning methods to handle this evolution grows. An indicative example is the single-cell RNA-seq (scRNA-seq), an emerging DNA sequencing technology with promising capabilities but significant computational challenges due to the large-scaled generated data. Regarding the classification process for scRNA-seq data, an appropriate method is the k Nearest Neighbor (kNN) classifier since it is usually utilized for large-scale prediction tasks due to its simplicity, minimal parameterization, and model-free nature. However, the ultra-high dimensionality that characterizes scRNA-seq impose a computational bottleneck, while prediction power can be affected by the "Curse of Dimensionality". In this work, we proposed the utilization of approximate nearest neighbor search algorithms for the task of kNN classification in scRNA-seq data focusing on a particular methodology tailored for high dimensional data. We argue that even relaxed approximate solutions will not affect the prediction performance significantly. The experimental results confirm the original assumption by offering the potential for broader applicability.
Real Time Sentiment Change Detection of Twitter Data Streams
Apr 02, 2018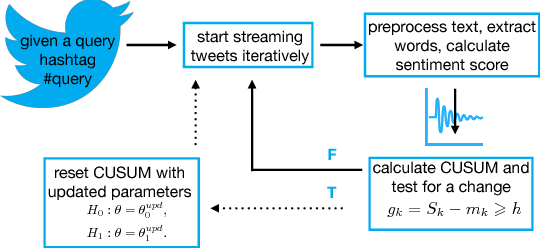

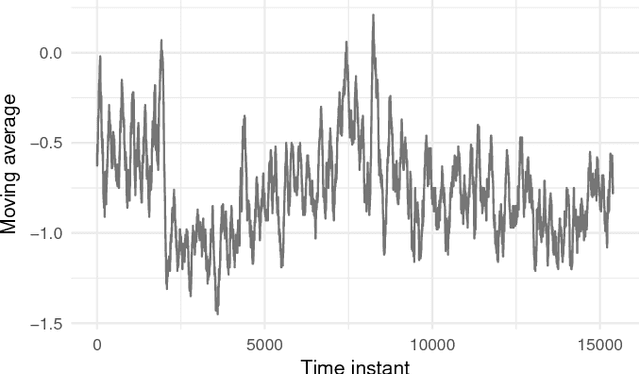
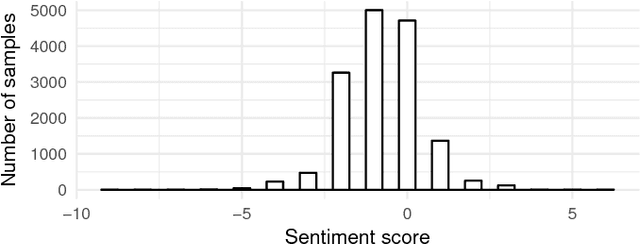
Abstract:In the past few years, there has been a huge growth in Twitter sentiment analysis having already provided a fair amount of research on sentiment detection of public opinion among Twitter users. Given the fact that Twitter messages are generated constantly with dizzying rates, a huge volume of streaming data is created, thus there is an imperative need for accurate methods for knowledge discovery and mining of this information. Although there exists a plethora of twitter sentiment analysis methods in the recent literature, the researchers have shifted to real-time sentiment identification on twitter streaming data, as expected. A major challenge is to deal with the Big Data challenges arising in Twitter streaming applications concerning both Volume and Velocity. Under this perspective, in this paper, a methodological approach based on open source tools is provided for real-time detection of changes in sentiment that is ultra efficient with respect to both memory consumption and computational cost. This is achieved by iteratively collecting tweets in real time and discarding them immediately after their process. For this purpose, we employ the Lexicon approach for sentiment characterizations, while change detection is achieved through appropriate control charts that do not require historical information. We believe that the proposed methodology provides the trigger for a potential large-scale monitoring of threads in an attempt to discover fake news spread or propaganda efforts in their early stages. Our experimental real-time analysis based on a recent hashtag provides evidence that the proposed approach can detect meaningful sentiment changes across a hashtags lifetime.
Convolutional Neural Networks for Toxic Comment Classification
Feb 27, 2018



Abstract:Flood of information is produced in a daily basis through the global Internet usage arising from the on-line interactive communications among users. While this situation contributes significantly to the quality of human life, unfortunately it involves enormous dangers, since on-line texts with high toxicity can cause personal attacks, on-line harassment and bullying behaviors. This has triggered both industrial and research community in the last few years while there are several tries to identify an efficient model for on-line toxic comment prediction. However, these steps are still in their infancy and new approaches and frameworks are required. On parallel, the data explosion that appears constantly, makes the construction of new machine learning computational tools for managing this information, an imperative need. Thankfully advances in hardware, cloud computing and big data management allow the development of Deep Learning approaches appearing very promising performance so far. For text classification in particular the use of Convolutional Neural Networks (CNN) have recently been proposed approaching text analytics in a modern manner emphasizing in the structure of words in a document. In this work, we employ this approach to discover toxic comments in a large pool of documents provided by a current Kaggle's competition regarding Wikipedia's talk page edits. To justify this decision we choose to compare CNNs against the traditional bag-of-words approach for text analysis combined with a selection of algorithms proven to be very effective in text classification. The reported results provide enough evidence that CNN enhance toxic comment classification reinforcing research interest towards this direction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge