Mohammad Junayed Hasan
CQ CNN: A Hybrid Classical Quantum Convolutional Neural Network for Alzheimer's Disease Detection Using Diffusion Generated and U Net Segmented 3D MRI
Mar 04, 2025Abstract:The detection of Alzheimer disease (AD) from clinical MRI data is an active area of research in medical imaging. Recent advances in quantum computing, particularly the integration of parameterized quantum circuits (PQCs) with classical machine learning architectures, offer new opportunities to develop models that may outperform traditional methods. However, quantum machine learning (QML) remains in its early stages and requires further experimental analysis to better understand its behavior and limitations. In this paper, we propose an end to end hybrid classical quantum convolutional neural network (CQ CNN) for AD detection using clinically formatted 3D MRI data. Our approach involves developing a framework to make 3D MRI data usable for machine learning, designing and training a brain tissue segmentation model (Skull Net), and training a diffusion model to generate synthetic images for the minority class. Our converged models exhibit potential quantum advantages, achieving higher accuracy in fewer epochs than classical models. The proposed beta8 3 qubit model achieves an accuracy of 97.50%, surpassing state of the art (SOTA) models while requiring significantly fewer computational resources. In particular, the architecture employs only 13K parameters (0.48 MB), reducing the parameter count by more than 99.99% compared to current SOTA models. Furthermore, the diffusion-generated data used to train our quantum models, in conjunction with real samples, preserve clinical structural standards, representing a notable first in the field of QML. We conclude that CQCNN architecture like models, with further improvements in gradient optimization techniques, could become a viable option and even a potential alternative to classical models for AD detection, especially in data limited and resource constrained clinical settings.
Shadow: A Novel Loss Function for Efficient Training in Siamese Networks
Nov 23, 2023Abstract:Despite significant recent advances in similarity detection tasks, existing approaches pose substantial challenges under memory constraints. One of the primary reasons for this is the use of computationally expensive metric learning loss functions such as Triplet Loss in Siamese networks. In this paper, we present a novel loss function called Shadow Loss that compresses the dimensions of an embedding space during loss calculation without loss of performance. The distance between the projections of the embeddings is learned from inputs on a compact projection space where distances directly correspond to a measure of class similarity. Projecting on a lower-dimension projection space, our loss function converges faster, and the resulting classified image clusters have higher inter-class and smaller intra-class distances. Shadow Loss not only reduces embedding dimensions favoring memory constraint devices but also consistently performs better than the state-of-the-art Triplet Margin Loss by an accuracy of 5\%-10\% across diverse datasets. The proposed loss function is also model agnostic, upholding its performance across several tested models. Its effectiveness and robustness across balanced, imbalanced, medical, and non-medical image datasets suggests that it is not specific to a particular model or dataset but demonstrates superior performance consistently while using less memory and computation.
Bridging Classical and Quantum Machine Learning: Knowledge Transfer From Classical to Quantum Neural Networks Using Knowledge Distillation
Nov 23, 2023Abstract:Very recently, studies have shown that quantum neural networks surpass classical neural networks in tasks like image classification when a similar number of learnable parameters are used. However, the development and optimization of quantum models are currently hindered by issues such as qubit instability and limited qubit availability, leading to error-prone systems with weak performance. In contrast, classical models can exhibit high-performance owing to substantial resource availability. As a result, more studies have been focusing on hybrid classical-quantum integration. A line of research particularly focuses on transfer learning through classical-quantum integration or quantum-quantum approaches. Unlike previous studies, this paper introduces a new method to transfer knowledge from classical to quantum neural networks using knowledge distillation, effectively bridging the gap between classical machine learning and emergent quantum computing techniques. We adapt classical convolutional neural network (CNN) architectures like LeNet and AlexNet to serve as teacher networks, facilitating the training of student quantum models by sending supervisory signals during backpropagation through KL-divergence. The approach yields significant performance improvements for the quantum models by solely depending on classical CNNs, with quantum models achieving an average accuracy improvement of 0.80% on the MNIST dataset and 5.40% on the more complex Fashion MNIST dataset. Applying this technique eliminates the cumbersome training of huge quantum models for transfer learning in resource-constrained settings and enables re-using existing pre-trained classical models to improve performance.Thus, this study paves the way for future research in quantum machine learning (QML) by positioning knowledge distillation as a core technique for advancing QML applications.
Preserving the knowledge of long clinical texts using aggregated ensembles of large language models
Nov 02, 2023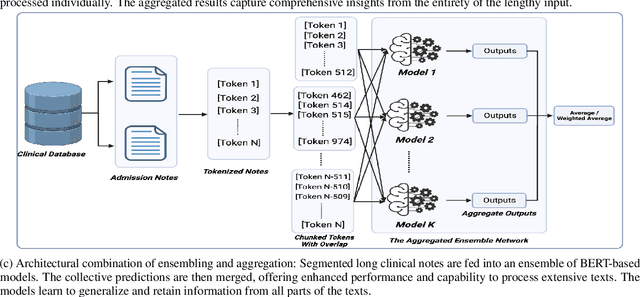
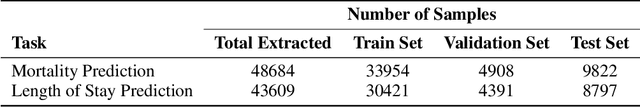
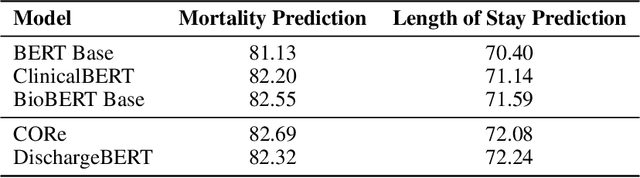
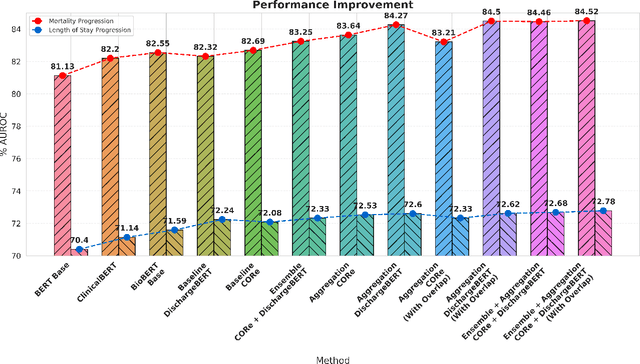
Abstract:Clinical texts, such as admission notes, discharge summaries, and progress notes, contain rich and valuable information that can be used for various clinical outcome prediction tasks. However, applying large language models, such as BERT-based models, to clinical texts poses two major challenges: the limitation of input length and the diversity of data sources. This paper proposes a novel method to preserve the knowledge of long clinical texts using aggregated ensembles of large language models. Unlike previous studies which use model ensembling or text aggregation methods separately, we combine ensemble learning with text aggregation and train multiple large language models on two clinical outcome tasks: mortality prediction and length of stay prediction. We show that our method can achieve better results than baselines, ensembling, and aggregation individually, and can improve the performance of large language models while handling long inputs and diverse datasets. We conduct extensive experiments on the admission notes from the MIMIC-III clinical database by combining multiple unstructured and high-dimensional datasets, demonstrating our method's effectiveness and superiority over existing approaches. We also provide a comprehensive analysis and discussion of our results, highlighting our method's applications and limitations for future research in the domain of clinical healthcare. The results and analysis of this study is supportive of our method assisting in clinical healthcare systems by enabling clinical decision-making with robust performance overcoming the challenges of long text inputs and varied datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge