Mobina Mansoori
Advancements in Medical Image Classification through Fine-Tuning Natural Domain Foundation Models
May 26, 2025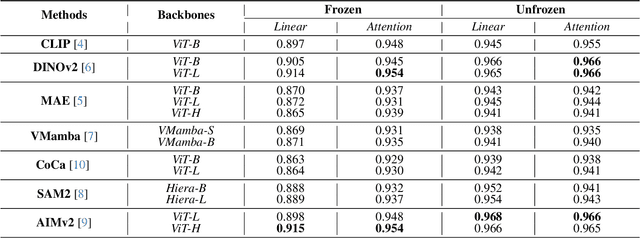
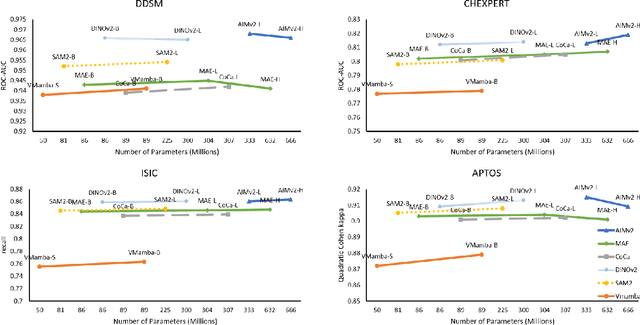
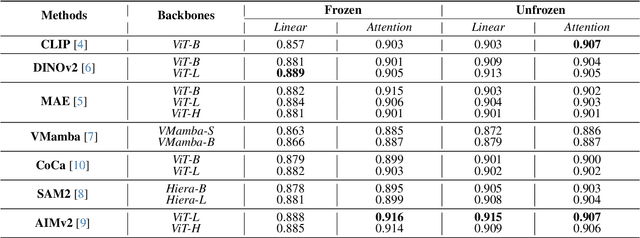
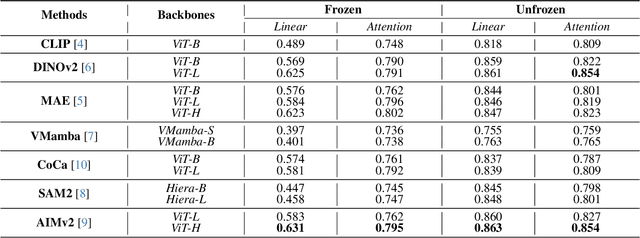
Abstract:Using massive datasets, foundation models are large-scale, pre-trained models that perform a wide range of tasks. These models have shown consistently improved results with the introduction of new methods. It is crucial to analyze how these trends impact the medical field and determine whether these advancements can drive meaningful change. This study investigates the application of recent state-of-the-art foundation models, DINOv2, MAE, VMamba, CoCa, SAM2, and AIMv2, for medical image classification. We explore their effectiveness on datasets including CBIS-DDSM for mammography, ISIC2019 for skin lesions, APTOS2019 for diabetic retinopathy, and CHEXPERT for chest radiographs. By fine-tuning these models and evaluating their configurations, we aim to understand the potential of these advancements in medical image classification. The results indicate that these advanced models significantly enhance classification outcomes, demonstrating robust performance despite limited labeled data. Based on our results, AIMv2, DINOv2, and SAM2 models outperformed others, demonstrating that progress in natural domain training has positively impacted the medical domain and improved classification outcomes. Our code is publicly available at: https://github.com/sajjad-sh33/Medical-Transfer-Learning.
The Missing Point in Vision Transformers for Universal Image Segmentation
May 26, 2025Abstract:Image segmentation remains a challenging task in computer vision, demanding robust mask generation and precise classification. Recent mask-based approaches yield high-quality masks by capturing global context. However, accurately classifying these masks, especially in the presence of ambiguous boundaries and imbalanced class distributions, remains an open challenge. In this work, we introduce ViT-P, a novel two-stage segmentation framework that decouples mask generation from classification. The first stage employs a proposal generator to produce class-agnostic mask proposals, while the second stage utilizes a point-based classification model built on the Vision Transformer (ViT) to refine predictions by focusing on mask central points. ViT-P serves as a pre-training-free adapter, allowing the integration of various pre-trained vision transformers without modifying their architecture, ensuring adaptability to dense prediction tasks. Furthermore, we demonstrate that coarse and bounding box annotations can effectively enhance classification without requiring additional training on fine annotation datasets, reducing annotation costs while maintaining strong performance. Extensive experiments across COCO, ADE20K, and Cityscapes datasets validate the effectiveness of ViT-P, achieving state-of-the-art results with 54.0 PQ on ADE20K panoptic segmentation, 87.4 mIoU on Cityscapes semantic segmentation, and 63.6 mIoU on ADE20K semantic segmentation. The code and pretrained models are available at: https://github.com/sajjad-sh33/ViT-P}{https://github.com/sajjad-sh33/ViT-P.
Self-Prompting Polyp Segmentation in Colonoscopy using Hybrid Yolo-SAM 2 Model
Sep 14, 2024



Abstract:Early diagnosis and treatment of polyps during colonoscopy are essential for reducing the incidence and mortality of Colorectal Cancer (CRC). However, the variability in polyp characteristics and the presence of artifacts in colonoscopy images and videos pose significant challenges for accurate and efficient polyp detection and segmentation. This paper presents a novel approach to polyp segmentation by integrating the Segment Anything Model (SAM 2) with the YOLOv8 model. Our method leverages YOLOv8's bounding box predictions to autonomously generate input prompts for SAM 2, thereby reducing the need for manual annotations. We conducted exhaustive tests on five benchmark colonoscopy image datasets and two colonoscopy video datasets, demonstrating that our method exceeds state-of-the-art models in both image and video segmentation tasks. Notably, our approach achieves high segmentation accuracy using only bounding box annotations, significantly reducing annotation time and effort. This advancement holds promise for enhancing the efficiency and scalability of polyp detection in clinical settings https://github.com/sajjad-sh33/YOLO_SAM2.
Polyp SAM 2: Advancing Zero shot Polyp Segmentation in Colorectal Cancer Detection
Aug 12, 2024



Abstract:Polyp segmentation plays a crucial role in the early detection and diagnosis of colorectal cancer. However, obtaining accurate segmentations often requires labor-intensive annotations and specialized models. Recently, Meta AI Research released a general Segment Anything Model 2 (SAM 2), which has demonstrated promising performance in several segmentation tasks. In this work, we evaluate the performance of SAM 2 in segmenting polyps under various prompted settings. We hope this report will provide insights to advance the field of polyp segmentation and promote more interesting work in the future. This project is publicly available at https://github.com/ sajjad-sh33/Polyp-SAM-2.
HistoSegCap: Capsules for Weakly-Supervised Semantic Segmentation of Histological Tissue Type in Whole Slide Images
Feb 16, 2024



Abstract:Digital pathology involves converting physical tissue slides into high-resolution Whole Slide Images (WSIs), which pathologists analyze for disease-affected tissues. However, large histology slides with numerous microscopic fields pose challenges for visual search. To aid pathologists, Computer Aided Diagnosis (CAD) systems offer visual assistance in efficiently examining WSIs and identifying diagnostically relevant regions. This paper presents a novel histopathological image analysis method employing Weakly Supervised Semantic Segmentation (WSSS) based on Capsule Networks, the first such application. The proposed model is evaluated using the Atlas of Digital Pathology (ADP) dataset and its performance is compared with other histopathological semantic segmentation methodologies. The findings underscore the potential of Capsule Networks in enhancing the precision and efficiency of histopathological image analysis. Experimental results show that the proposed model outperforms traditional methods in terms of accuracy and the mean Intersection-over-Union (mIoU) metric.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge