Mitsuhiro Nakamura
Construction of an Organ Shape Atlas Using a Hierarchical Mesh Variational Autoencoder
Jun 18, 2025Abstract:An organ shape atlas, which represents the shape and position of the organs and skeleton of a living body using a small number of parameters, is expected to have a wide range of clinical applications, including intraoperative guidance and radiotherapy. Because the shape and position of soft organs vary greatly among patients, it is difficult for linear models to reconstruct shapes that have large local variations. Because it is difficult for conventional nonlinear models to control and interpret the organ shapes obtained, deep learning has been attracting attention in three-dimensional shape representation. In this study, we propose an organ shape atlas based on a mesh variational autoencoder (MeshVAE) with hierarchical latent variables. To represent the complex shapes of biological organs and nonlinear shape differences between individuals, the proposed method maintains the performance of organ shape reconstruction by hierarchizing latent variables and enables shape representation using lower-dimensional latent variables. Additionally, templates that define vertex correspondence between different resolutions enable hierarchical representation in mesh data and control the global and local features of the organ shape. We trained the model using liver and stomach organ meshes obtained from 124 cases and confirmed that the model reconstructed the position and shape with an average distance between vertices of 1.5 mm and mean distance of 0.7 mm for the liver shape, and an average distance between vertices of 1.4 mm and mean distance of 0.8 mm for the stomach shape on test data from 19 of cases. The proposed method continuously represented interpolated shapes, and by changing latent variables at different hierarchical levels, the proposed method hierarchically separated shape features compared with PCA.
2D/3D Deep Image Registration by Learning 3D Displacement Fields for Abdominal Organs
Dec 11, 2022Abstract:Deformable registration of two-dimensional/three-dimensional (2D/3D) images of abdominal organs is a complicated task because the abdominal organs deform significantly and their contours are not detected in two-dimensional X-ray images. We propose a supervised deep learning framework that achieves 2D/3D deformable image registration between 3D volumes and single-viewpoint 2D projected images. The proposed method learns the translation from the target 2D projection images and the initial 3D volume to 3D displacement fields. In experiments, we registered 3D-computed tomography (CT) volumes to digitally reconstructed radiographs generated from abdominal 4D-CT volumes. For validation, we used 4D-CT volumes of 35 cases and confirmed that the 3D-CT volumes reflecting the nonlinear and local respiratory organ displacement were reconstructed. The proposed method demonstrate the compatible performance to the conventional methods with a dice similarity coefficient of 91.6 \% for the liver region and 85.9 \% for the stomach region, while estimating a significantly more accurate CT values.
Feedback Assisted Adversarial Learning to Improve the Quality of Cone-beam CT Images
Oct 23, 2022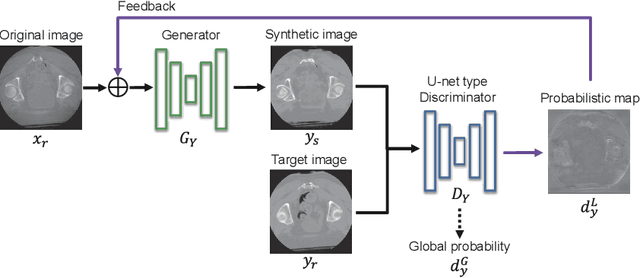

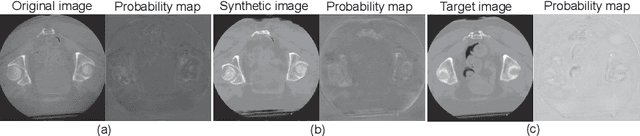
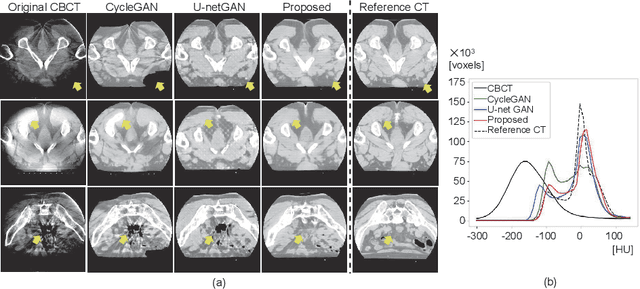
Abstract:Unsupervised image translation using adversarial learning has been attracting attention to improve the image quality of medical images. However, adversarial training based on the global evaluation values of discriminators does not provide sufficient translation performance for locally different image features. We propose adversarial learning with a feedback mechanism from a discriminator to improve the quality of CBCT images. This framework employs U-net as the discriminator and outputs a probability map representing the local discrimination results. The probability map is fed back to the generator and used for training to improve the image translation. Our experiments using 76 corresponding CT-CBCT images confirmed that the proposed framework could capture more diverse image features than conventional adversarial learning frameworks and produced synthetic images with pixel values close to the reference image and a correlation coefficient of 0.93.
IGCN: Image-to-graph Convolutional Network for 2D/3D Deformable Registration
Oct 31, 2021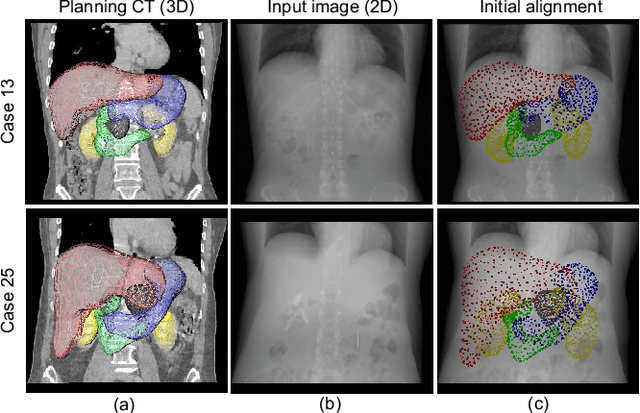
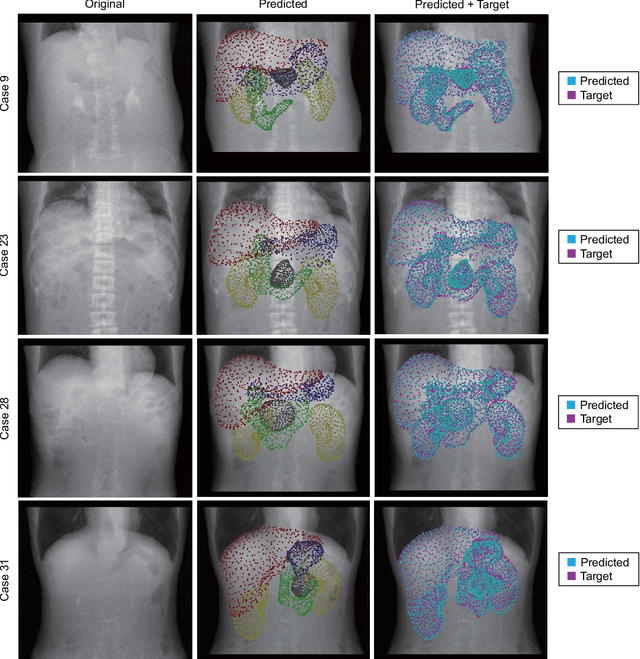
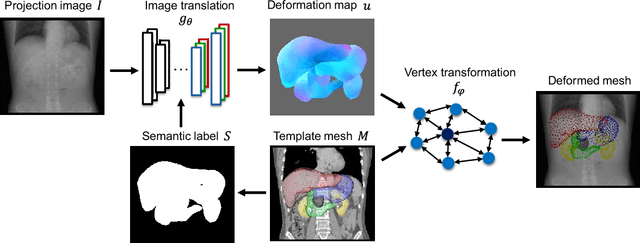
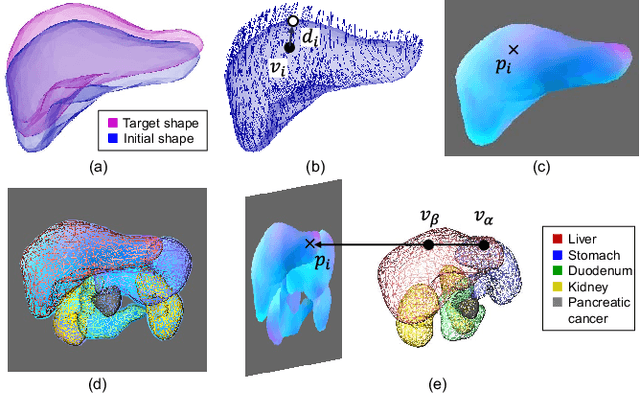
Abstract:Organ shape reconstruction based on a single-projection image during treatment has wide clinical scope, e.g., in image-guided radiotherapy and surgical guidance. We propose an image-to-graph convolutional network that achieves deformable registration of a 3D organ mesh for a single-viewpoint 2D projection image. This framework enables simultaneous training of two types of transformation: from the 2D projection image to a displacement map, and from the sampled per-vertex feature to a 3D displacement that satisfies the geometrical constraint of the mesh structure. Assuming application to radiation therapy, the 2D/3D deformable registration performance is verified for multiple abdominal organs that have not been targeted to date, i.e., the liver, stomach, duodenum, and kidney, and for pancreatic cancer. The experimental results show shape prediction considering relationships among multiple organs can be used to predict respiratory motion and deformation from digitally reconstructed radiographs with clinically acceptable accuracy.
Statistical Deformation Reconstruction Using Multi-organ Shape Features for Pancreatic Cancer Localization
Nov 13, 2019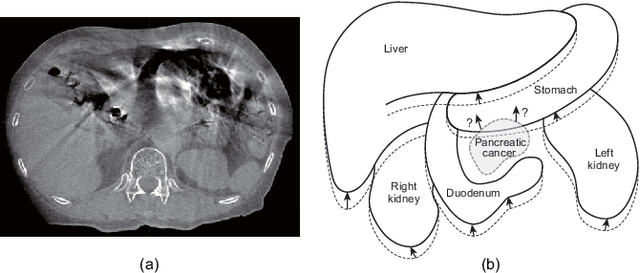
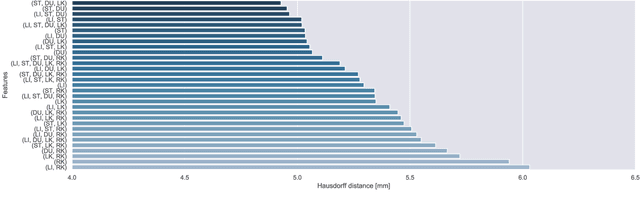
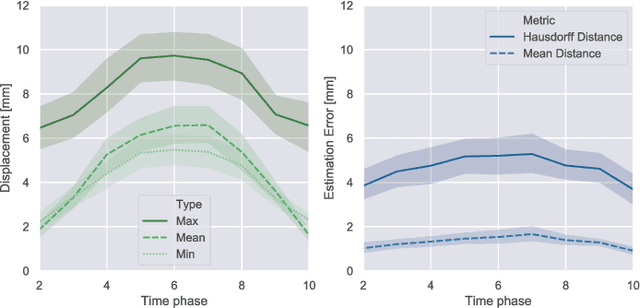
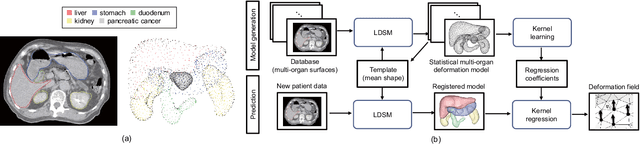
Abstract:Respiratory motion and the associated deformations of abdominal organs and tumors are essential information in clinical applications. However, inter- and intra-patient multi-organ deformations are complex and have not been statistically formulated, whereas single organ deformations have been widely studied. In this paper, we introduce a multi-organ deformation library and its application to deformation reconstruction based on the shape features of multiple abdominal organs. Statistical multi-organ motion/deformation models of the stomach, liver, left and right kidneys, and duodenum were generated by shape matching their region labels defined on four-dimensional computed tomography images. A total of 250 volumes were measured from 25 pancreatic cancer patients. This paper also proposes a per-region-based deformation learning using the reproducing kernel to predict the displacement of pancreatic cancer for adaptive radiotherapy. The experimental results show that the proposed concept estimates deformations better than general per-patient-based learning models and achieves a clinically acceptable estimation error with a mean distance of 1.2 $\pm$ 0.7 mm and a Hausdorff distance of 4.2 $\pm$ 2.3 mm throughout the respiratory motion.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge