Miguel Morales
Neuron detection in stack images: a persistent homology interpretation
Sep 15, 2015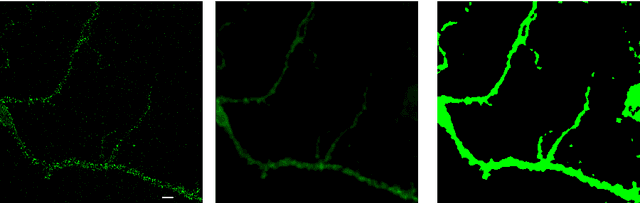
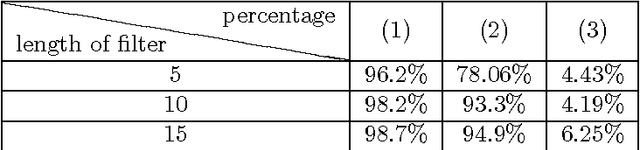
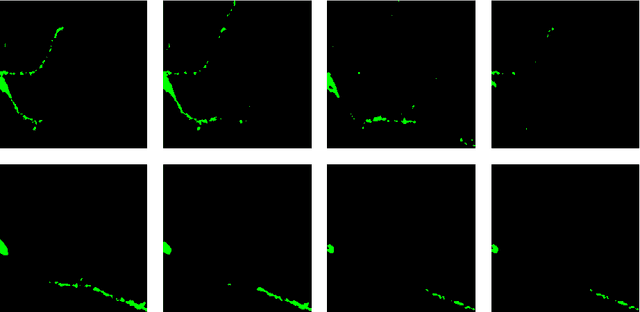
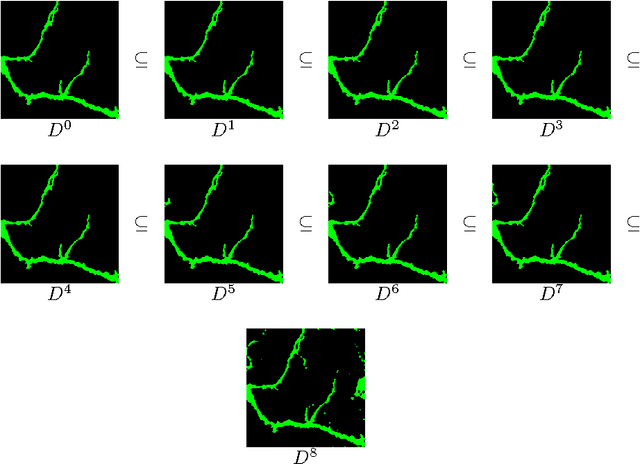
Abstract:Automation and reliability are the two main requirements when computers are applied in Life Sciences. In this paper we report on an application to neuron recognition, an important step in our long-term project of providing software systems to the study of neural morphology and functionality from biomedical images. Our algorithms have been implemented in an ImageJ plugin called NeuronPersistentJ, which has been validated experimentally. The soundness and reliability of our approach are based on the interpretation of our processing methods with respect to persistent homology, a well-known tool in computational mathematics.
SynapCountJ --- a Tool for Analyzing Synaptic Densities in Neurons
Jul 28, 2015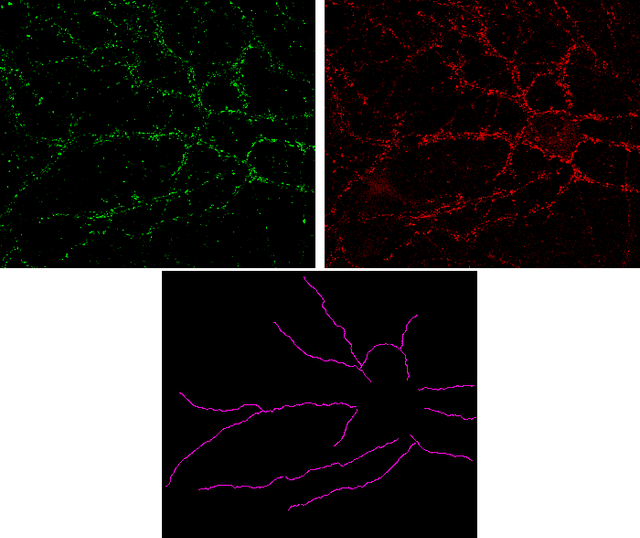
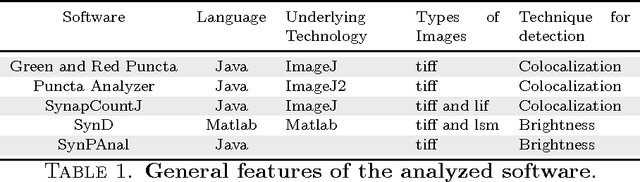
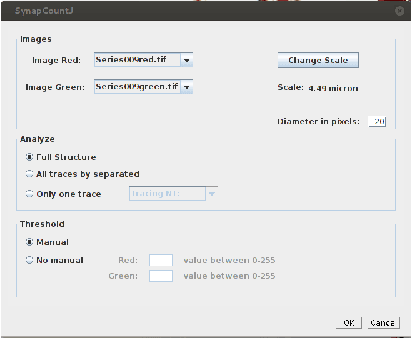
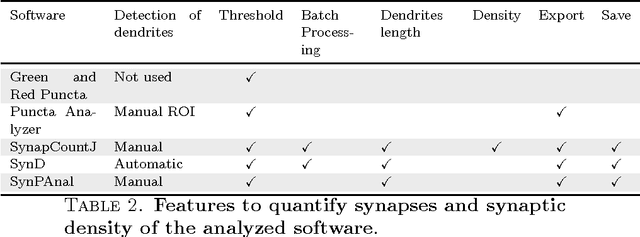
Abstract:The quantification of synapses is instrumental to measure the evolution of synaptic densities of neurons under the effect of some physiological conditions, neuronal diseases or even drug treatments. However, the manual quantification of synapses is a tedious, error-prone, time-consuming and subjective task; therefore, tools that might automate this process are desirable. In this paper, we present SynapCountJ, an ImageJ plugin, that can measure synaptic density of individual neurons obtained by immunofluorescence techniques, and also can be applied for batch processing of neurons that have been obtained in the same experiment or using the same setting. The procedure to quantify synapses implemented in SynapCountJ is based on the colocalization of three images of the same neuron (the neuron marked with two antibody markers and the structure of the neuron) and is inspired by methods coming from Computational Algebraic Topology. SynapCountJ provides a procedure to semi-automatically quantify the number of synapses of neuron cultures; as a result, the time required for such an analysis is greatly reduced.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge