Michele Monti
Prediction of gene expression time series and structural analysis of gene regulatory networks using recurrent neural networks
Sep 13, 2021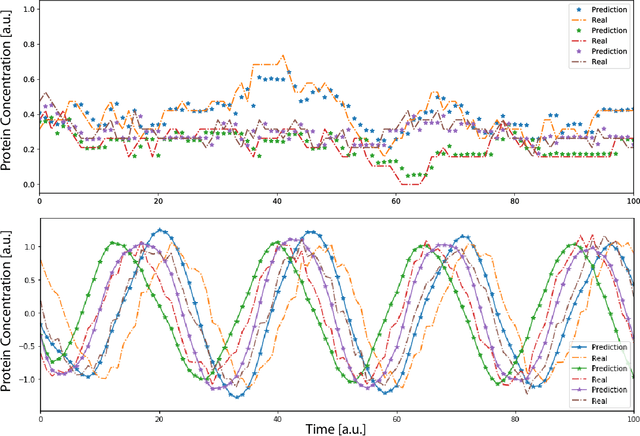
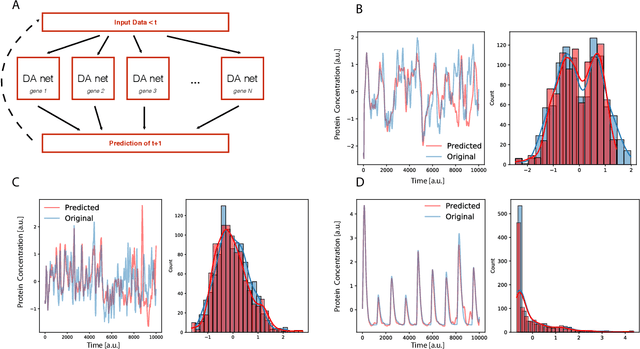
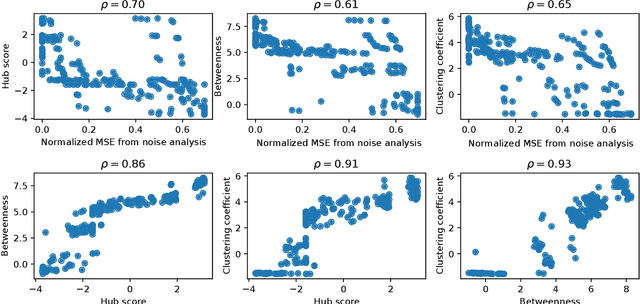
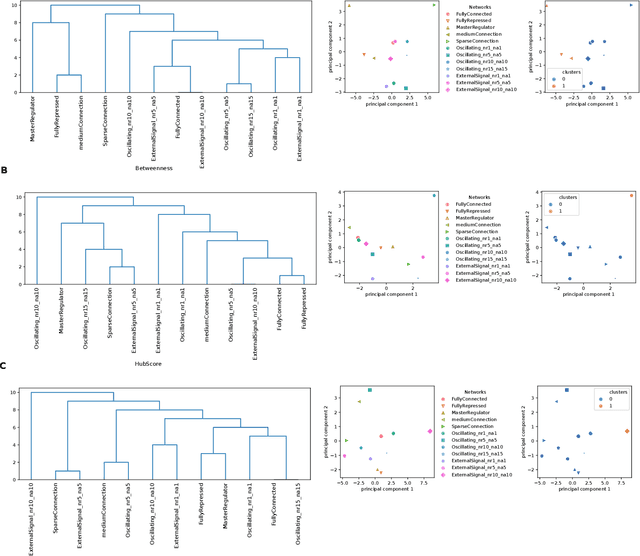
Abstract:Methods for time series prediction and classification of gene regulatory networks (GRNs) from gene expression data have been treated separately so far. The recent emergence of attention-based recurrent neural networks (RNN) models boosted the interpretability of RNN parameters, making them appealing for the understanding of gene interactions. In this work, we generated synthetic time series gene expression data from a range of archetypal GRNs and we relied on a dual attention RNN to predict the gene temporal dynamics. We show that the prediction is extremely accurate for GRNs with different architectures. Next, we focused on the attention mechanism of the RNN and, using tools from graph theory, we found that its graph properties allow to hierarchically distinguish different architectures of the GRN. We show that the GRNs respond differently to the addition of noise in the prediction by the RNN and we relate the noise response to the analysis of the attention mechanism. In conclusion, this work provides a a way to understand and exploit the attention mechanism of RNN and it paves the way to RNN-based methods for time series prediction and inference of GRNs from gene expression data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge