Giorgio Gosti
Evidence of Scaling Regimes in the Hopfield Dynamics of Whole Brain Model
Jan 15, 2024Abstract:It is shown that a Hopfield recurrent neural network, informed by experimentally derived brain topology, recovers the scaling picture recently introduced by Deco et al., according to which the process of information transfer within the human brain shows spatially correlated patterns qualitatively similar to those displayed by turbulent flows. Although both models employ a coupling strength which decays exponentially with the euclidean distance between the nodes, their mathematical nature is widely different, Hopf oscillators versus Hopfield neural network. Hence, their convergence suggests a remarkable robustness of the aforementioned scaling picture. Furthermore, the present analysis shows that the Hopfield model brain remains functional by removing links above about five decay lengths, corresponding to about one sixth of the size of the global brain. This suggests that, in terms of connectivity decay length, the Hopfield brain functions in a sort of intermediate "turbulent liquid"-like state, whose essential connections are the intermediate ones between the connectivity decay length and the global brain size. This "turbulent-like liquid" appears to be more spiky than actual turbulent fluids, with a scaling exponent around $2/5$ instead of $2/3$.
Prediction of gene expression time series and structural analysis of gene regulatory networks using recurrent neural networks
Sep 13, 2021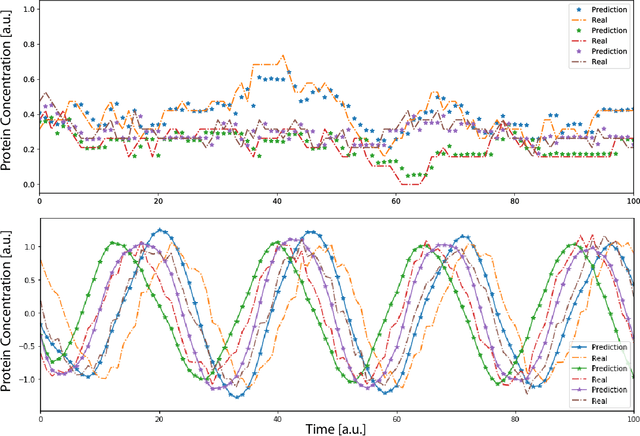
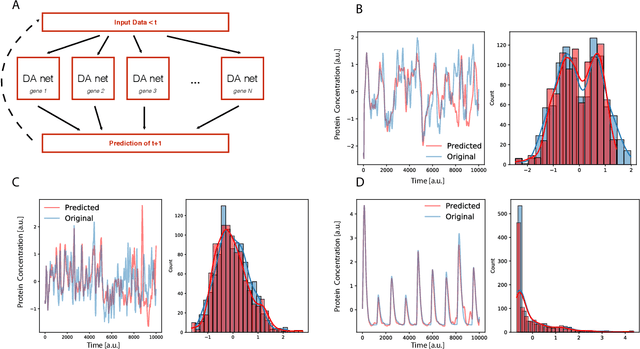
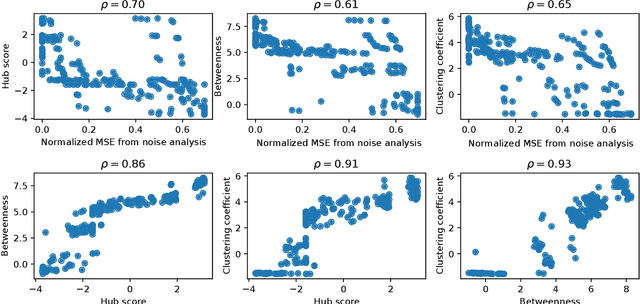
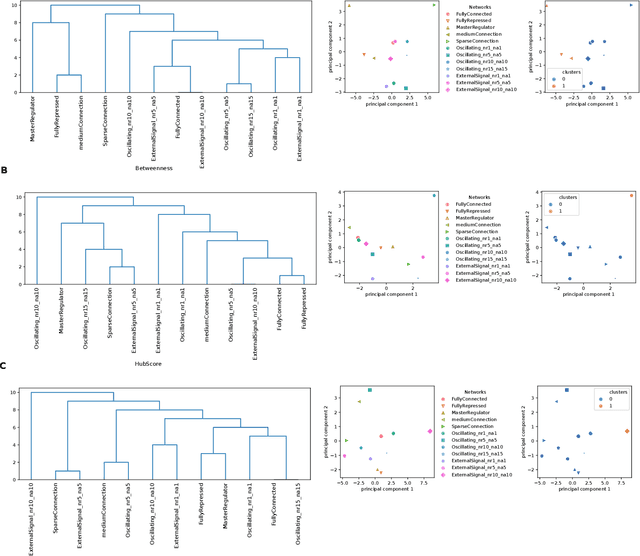
Abstract:Methods for time series prediction and classification of gene regulatory networks (GRNs) from gene expression data have been treated separately so far. The recent emergence of attention-based recurrent neural networks (RNN) models boosted the interpretability of RNN parameters, making them appealing for the understanding of gene interactions. In this work, we generated synthetic time series gene expression data from a range of archetypal GRNs and we relied on a dual attention RNN to predict the gene temporal dynamics. We show that the prediction is extremely accurate for GRNs with different architectures. Next, we focused on the attention mechanism of the RNN and, using tools from graph theory, we found that its graph properties allow to hierarchically distinguish different architectures of the GRN. We show that the GRNs respond differently to the addition of noise in the prediction by the RNN and we relate the noise response to the analysis of the attention mechanism. In conclusion, this work provides a a way to understand and exploit the attention mechanism of RNN and it paves the way to RNN-based methods for time series prediction and inference of GRNs from gene expression data.
Effect of dilution in asymmetric recurrent neural networks
May 10, 2018



Abstract:We study with numerical simulation the possible limit behaviors of synchronous discrete-time deterministic recurrent neural networks composed of N binary neurons as a function of a network's level of dilution and asymmetry. The network dilution measures the fraction of neuron couples that are connected, and the network asymmetry measures to what extent the underlying connectivity matrix is asymmetric. For each given neural network, we study the dynamical evolution of all the different initial conditions, thus characterizing the full dynamical landscape without imposing any learning rule. Because of the deterministic dynamics, each trajectory converges to an attractor, that can be either a fixed point or a limit cycle. These attractors form the set of all the possible limit behaviors of the neural network. For each network, we then determine the convergence times, the limit cycles' length, the number of attractors, and the sizes of the attractors' basin. We show that there are two network structures that maximize the number of possible limit behaviors. The first optimal network structure is fully-connected and symmetric. On the contrary, the second optimal network structure is highly sparse and asymmetric. The latter optimal is similar to what observed in different biological neuronal circuits. These observations lead us to hypothesize that independently from any given learning model, an efficient and effective biologic network that stores a number of limit behaviors close to its maximum capacity tends to develop a connectivity structure similar to one of the optimal networks we found.
* 31 pages, 5 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge