Michael Betenbaugh
Data-driven and Physics Informed Modelling of Chinese Hamster Ovary Cell Bioreactors
May 05, 2023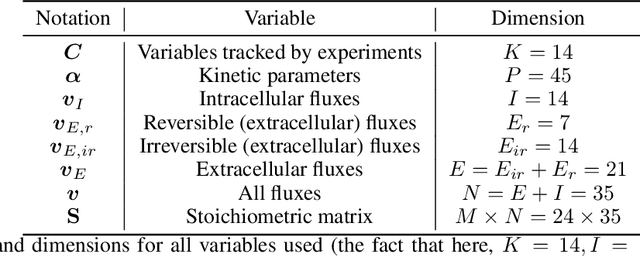



Abstract:Fed-batch culture is an established operation mode for the production of biologics using mammalian cell cultures. Quantitative modeling integrates both kinetics for some key reaction steps and optimization-driven metabolic flux allocation, using flux balance analysis; this is known to lead to certain mathematical inconsistencies. Here, we propose a physically-informed data-driven hybrid model (a "gray box") to learn models of the dynamical evolution of Chinese Hamster Ovary (CHO) cell bioreactors from process data. The approach incorporates physical laws (e.g. mass balances) as well as kinetic expressions for metabolic fluxes. Machine learning (ML) is then used to (a) directly learn evolution equations (black-box modelling); (b) recover unknown physical parameters ("white-box" parameter fitting) or -- importantly -- (c) learn partially unknown kinetic expressions (gray-box modelling). We encode the convex optimization step of the overdetermined metabolic biophysical system as a differentiable, feed-forward layer into our architectures, connecting partial physical knowledge with data-driven machine learning.
Some of the variables, some of the parameters, some of the times, with some physics known: Identification with partial information
Apr 27, 2023Abstract:Experimental data is often comprised of variables measured independently, at different sampling rates (non-uniform ${\Delta}$t between successive measurements); and at a specific time point only a subset of all variables may be sampled. Approaches to identifying dynamical systems from such data typically use interpolation, imputation or subsampling to reorganize or modify the training data $\textit{prior}$ to learning. Partial physical knowledge may also be available $\textit{a priori}$ (accurately or approximately), and data-driven techniques can complement this knowledge. Here we exploit neural network architectures based on numerical integration methods and $\textit{a priori}$ physical knowledge to identify the right-hand side of the underlying governing differential equations. Iterates of such neural-network models allow for learning from data sampled at arbitrary time points $\textit{without}$ data modification. Importantly, we integrate the network with available partial physical knowledge in "physics informed gray-boxes"; this enables learning unknown kinetic rates or microbial growth functions while simultaneously estimating experimental parameters.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge