Mert Aral
Development of an accessible 10-year Digital CArdioVAscular risk assessment: a UK Biobank study
Apr 20, 2021
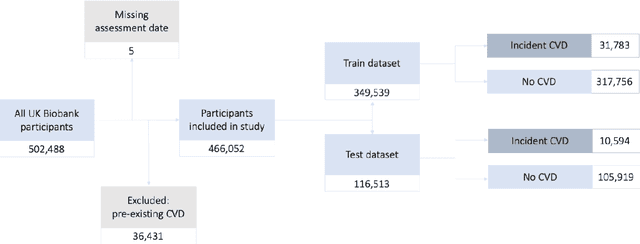
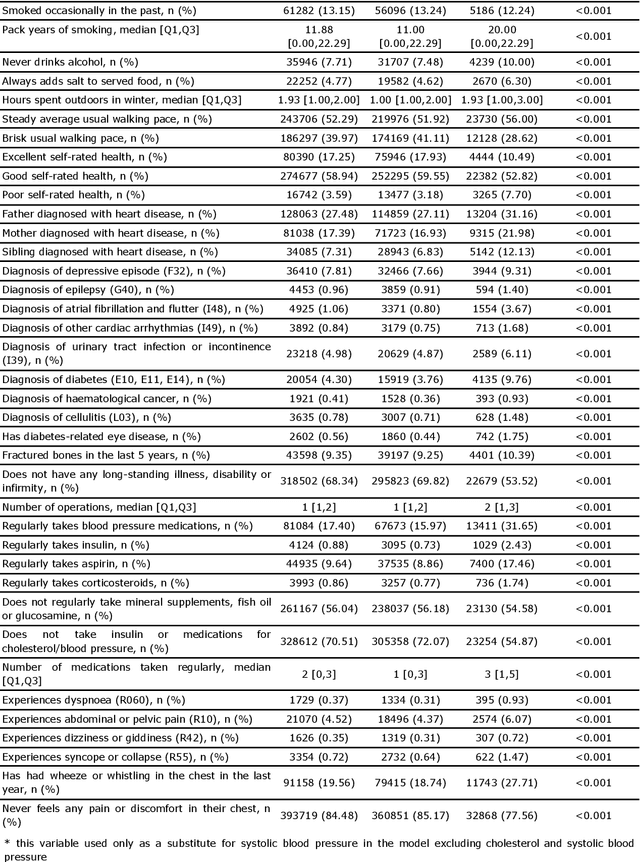
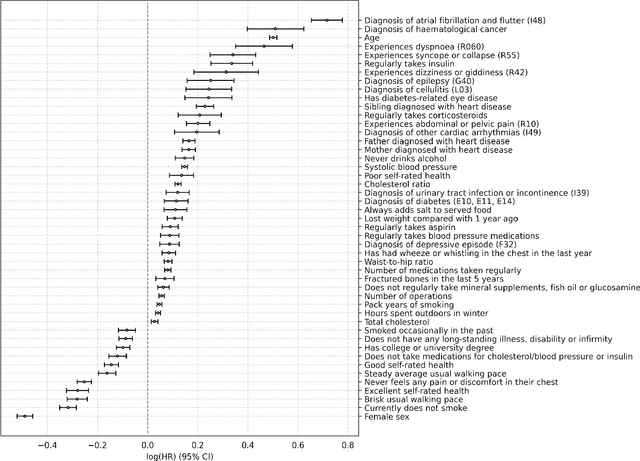
Abstract:Background: Cardiovascular diseases (CVDs) are among the leading causes of death worldwide. Predictive scores providing personalised risk of developing CVD are increasingly used in clinical practice. Most scores, however, utilise a homogenous set of features and require the presence of a physician. Objective: The aim was to develop a new risk model (DiCAVA) using statistical and machine learning techniques that could be applied in a remote setting. A secondary goal was to identify new patient-centric variables that could be incorporated into CVD risk assessments. Methods: Across 466,052 participants, Cox proportional hazards (CPH) and DeepSurv models were trained using 608 variables derived from the UK Biobank to investigate the 10-year risk of developing a CVD. Data-driven feature selection reduced the number of features to 47, after which reduced models were trained. Both models were compared to the Framingham score. Results: The reduced CPH model achieved a c-index of 0.7443, whereas DeepSurv achieved a c-index of 0.7446. Both CPH and DeepSurv were superior in determining the CVD risk compared to Framingham score. Minimal difference was observed when cholesterol and blood pressure were excluded from the models (CPH: 0.741, DeepSurv: 0.739). The models show very good calibration and discrimination on the test data. Conclusion: We developed a cardiovascular risk model that has very good predictive capacity and encompasses new variables. The score could be incorporated into clinical practice and utilised in a remote setting, without the need of including cholesterol. Future studies will focus on external validation across heterogeneous samples.
Machine learning approach to dynamic risk modeling of mortality in COVID-19: a UK Biobank study
Apr 19, 2021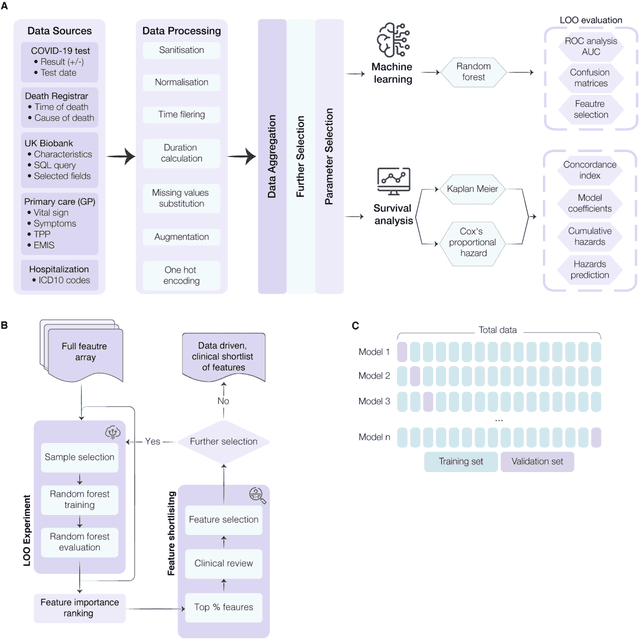
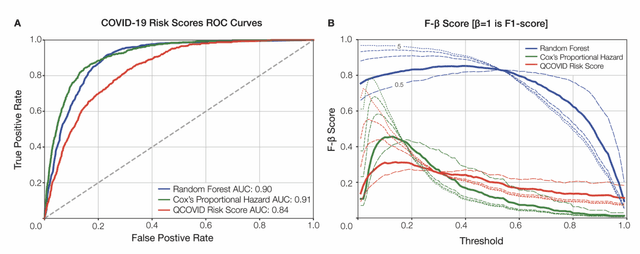
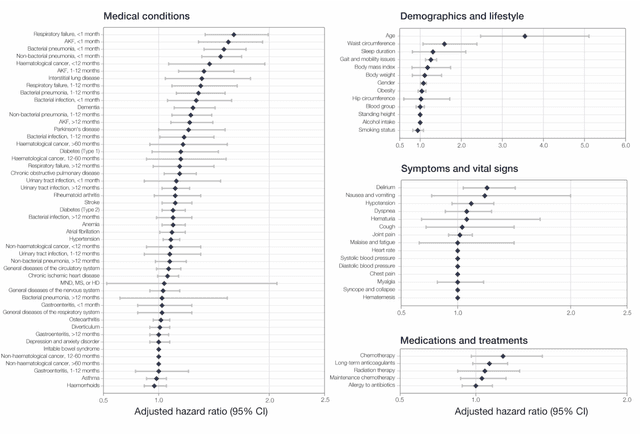
Abstract:The COVID-19 pandemic has created an urgent need for robust, scalable monitoring tools supporting stratification of high-risk patients. This research aims to develop and validate prediction models, using the UK Biobank, to estimate COVID-19 mortality risk in confirmed cases. From the 11,245 participants testing positive for COVID-19, we develop a data-driven random forest classification model with excellent performance (AUC: 0.91), using baseline characteristics, pre-existing conditions, symptoms, and vital signs, such that the score could dynamically assess mortality risk with disease deterioration. We also identify several significant novel predictors of COVID-19 mortality with equivalent or greater predictive value than established high-risk comorbidities, such as detailed anthropometrics and prior acute kidney failure, urinary tract infection, and pneumonias. The model design and feature selection enables utility in outpatient settings. Possible applications include supporting individual-level risk profiling and monitoring disease progression across patients with COVID-19 at-scale, especially in hospital-at-home settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge