Md. Muhtadir Rahman
An Ensemble-based Multi-Criteria Decision Making Method for COVID-19 Cough Classification
Oct 01, 2021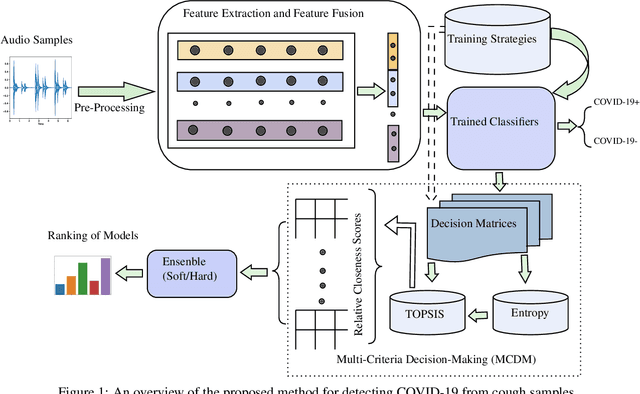
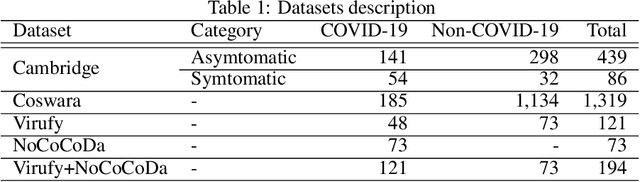
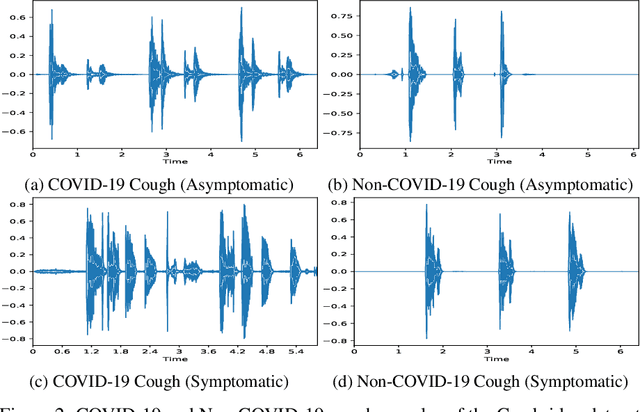
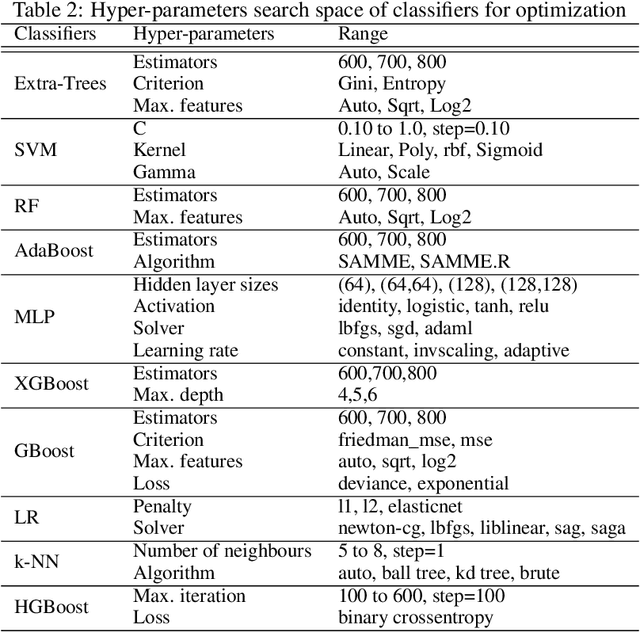
Abstract:The objectives of this research are analysing the performance of the state-of-the-art machine learning techniques for classifying COVID-19 from cough sound and identifying the model(s) that consistently perform well across different cough datasets. Different performance evaluation metrics (such as precision, sensitivity, specificity, AUC, accuracy, etc.) make it difficult to select the best performance model. To address this issue, in this paper, we propose an ensemble-based multi-criteria decision making (MCDM) method for selecting top performance machine learning technique(s) for COVID-19 cough classification. We use four cough datasets, namely Cambridge, Coswara, Virufy, and NoCoCoDa to verify the proposed method. At first, our proposed method uses the audio features of cough samples and then applies machine learning (ML) techniques to classify them as COVID-19 or non-COVID-19. Then, we consider a multi-criteria decision-making (MCDM) method that combines ensemble technologies (i.e., soft and hard) to select the best model. In MCDM, we use the technique for order preference by similarity to ideal solution (TOPSIS) for ranking purposes, while entropy is applied to calculate evaluation criteria weights. In addition, we apply the feature reduction process through recursive feature elimination with cross-validation under different estimators. The results of our empirical evaluations show that the proposed method outperforms the state-of-the-art models.
ECOVNet: An Ensemble of Deep Convolutional Neural Networks Based on EfficientNet to Detect COVID-19 From Chest X-rays
Oct 16, 2020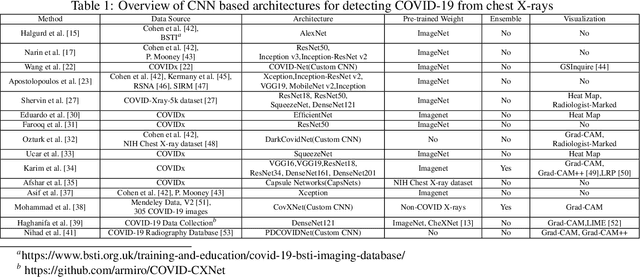
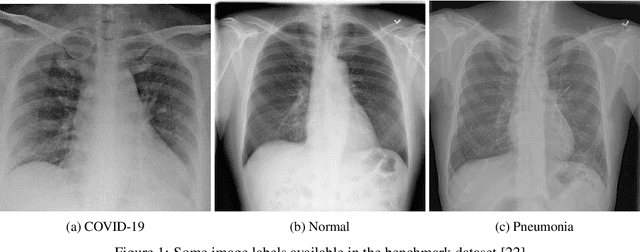

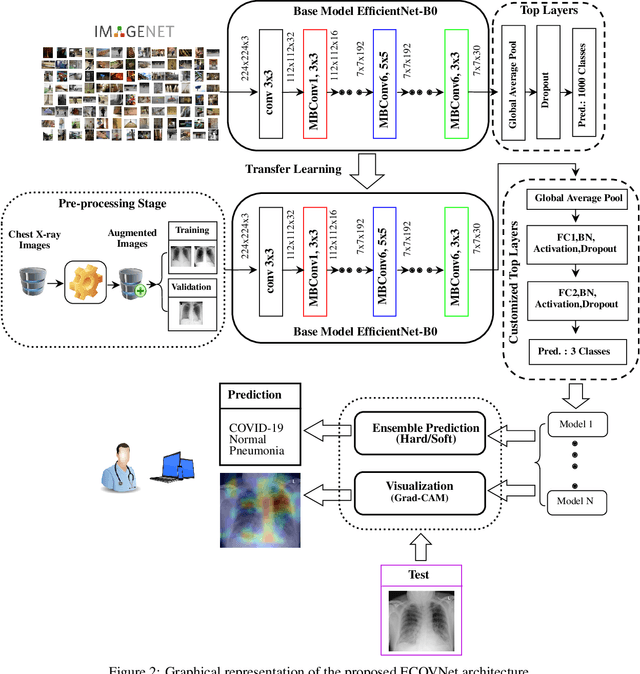
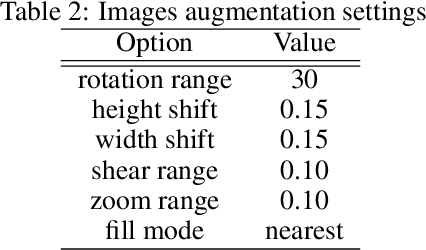
Abstract:This paper proposed an ensemble of deep convolutional neural networks (CNN) based on EfficientNet, named ECOVNet, to detect COVID-19 using a large chest X-ray data set. At first, the open-access large chest X-ray collection is augmented, and then ImageNet pre-trained weights for EfficientNet is transferred with some customized fine-tuning top layers that are trained, followed by an ensemble of model snapshots to classify chest X-rays corresponding to COVID-19, normal, and pneumonia. The predictions of the model snapshots, which are created during a single training, are combined through two ensemble strategies, i.e., hard ensemble and soft ensemble to ameliorate classification performance and generalization in the related task of classifying chest X-rays.
PDCOVIDNet: A Parallel-Dilated Convolutional Neural Network Architecture for Detecting COVID-19 from Chest X-Ray Images
Jul 29, 2020

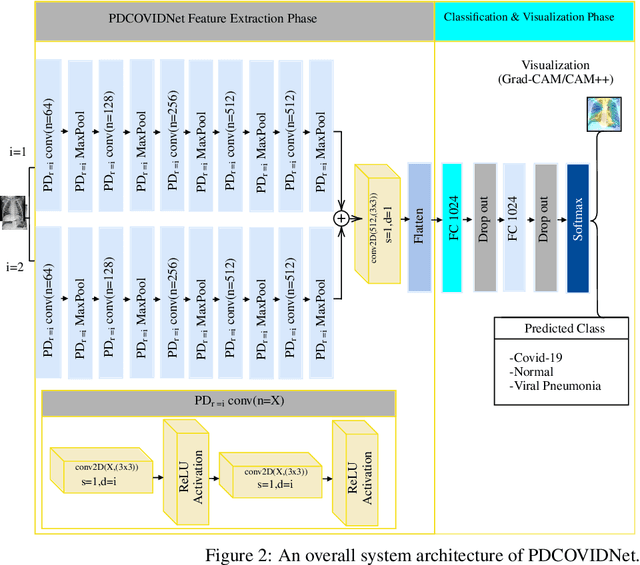
Abstract:The COVID-19 pandemic continues to severely undermine the prosperity of the global health system. To combat this pandemic, effective screening techniques for infected patients are indispensable. There is no doubt that the use of chest X-ray images for radiological assessment is one of the essential screening techniques. Some of the early studies revealed that the patient's chest X-ray images showed abnormalities, which is natural for patients infected with COVID-19. In this paper, we proposed a parallel-dilated convolutional neural network (CNN) based COVID-19 detection system from chest x-ray images, named as Parallel-Dilated COVIDNet (PDCOVIDNet). First, the publicly available chest X-ray collection fully preloaded and enhanced, and then classified by the proposed method. Differing convolution dilation rate in a parallel form demonstrates the proof-of-principle for using PDCOVIDNet to extract radiological features for COVID-19 detection. Accordingly, we have assisted our method with two visualization methods, which are specifically designed to increase understanding of the key components associated with COVID-19 infection. Both visualization methods compute gradients for a given image category related to feature maps of the last convolutional layer to create a class-discriminative region. In our experiment, we used a total of 2,905 chest X-ray images, comprising three cases (such as COVID-19, normal, and viral pneumonia), and empirical evaluations revealed that the proposed method extracted more significant features expeditiously related to the suspected disease. The experimental results demonstrate that our proposed method significantly improves performance metrics: accuracy, precision, recall, and F1 scores reach 96.58%, 96.58%, 96.59%, and 96.58%, respectively, which is comparable or enhanced compared with the state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge