Max Wasserman
Stabilizing the Kumaraswamy Distribution
Oct 01, 2024Abstract:Large-scale latent variable models require expressive continuous distributions that support efficient sampling and low-variance differentiation, achievable through the reparameterization trick. The Kumaraswamy (KS) distribution is both expressive and supports the reparameterization trick with a simple closed-form inverse CDF. Yet, its adoption remains limited. We identify and resolve numerical instabilities in the inverse CDF and log-pdf, exposing issues in libraries like PyTorch and TensorFlow. We then introduce simple and scalable latent variable models based on the KS, improving exploration-exploitation trade-offs in contextual multi-armed bandits and enhancing uncertainty quantification for link prediction with graph neural networks. Our results support the stabilized KS distribution as a core component in scalable variational models for bounded latent variables.
Graph Structure Learning with Interpretable Bayesian Neural Networks
Jun 20, 2024Abstract:Graphs serve as generic tools to encode the underlying relational structure of data. Often this graph is not given, and so the task of inferring it from nodal observations becomes important. Traditional approaches formulate a convex inverse problem with a smoothness promoting objective and rely on iterative methods to obtain a solution. In supervised settings where graph labels are available, one can unroll and truncate these iterations into a deep network that is trained end-to-end. Such a network is parameter efficient and inherits inductive bias from the optimization formulation, an appealing aspect for data constrained settings in, e.g., medicine, finance, and the natural sciences. But typically such settings care equally about uncertainty over edge predictions, not just point estimates. Here we introduce novel iterations with independently interpretable parameters, i.e., parameters whose values - independent of other parameters' settings - proportionally influence characteristics of the estimated graph, such as edge sparsity. After unrolling these iterations, prior knowledge over such graph characteristics shape prior distributions over these independently interpretable network parameters to yield a Bayesian neural network (BNN) capable of graph structure learning (GSL) from smooth signal observations. Fast execution and parameter efficiency allow for high-fidelity posterior approximation via Markov Chain Monte Carlo (MCMC) and thus uncertainty quantification on edge predictions. Synthetic and real data experiments corroborate this model's ability to provide well-calibrated estimates of uncertainty, in test cases that include unveiling economic sector modular structure from S$\&$P$500$ data and recovering pairwise digit similarities from MNIST images. Overall, this framework enables GSL in modest-scale applications where uncertainty on the data structure is paramount.
pyGSL: A Graph Structure Learning Toolkit
Nov 07, 2022


Abstract:We introduce pyGSL, a Python library that provides efficient implementations of state-of-the-art graph structure learning models along with diverse datasets to evaluate them on. The implementations are written in GPU-friendly ways, allowing one to scale to much larger network tasks. A common interface is introduced for algorithm unrolling methods, unifying implementations of recent state-of-the-art techniques and allowing new methods to be quickly developed by avoiding the need to rebuild the underlying unrolling infrastructure. Implementations of differentiable graph structure learning models are written in PyTorch, allowing us to leverage the rich software ecosystem that exists e.g., around logging, hyperparameter search, and GPU-communication. This also makes it easy to incorporate these models as components in larger gradient based learning systems where differentiable estimates of graph structure may be useful, e.g. in latent graph learning. Diverse datasets and performance metrics allow consistent comparisons across models in this fast growing field. The full code repository can be found on https://github.com/maxwass/pyGSL.
Learning Graph Structure from Convolutional Mixtures
May 19, 2022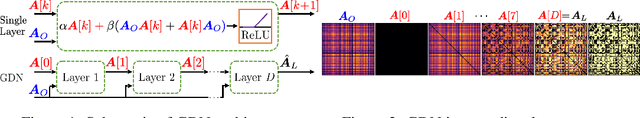

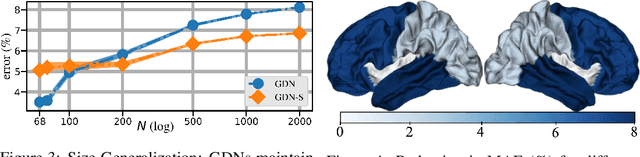
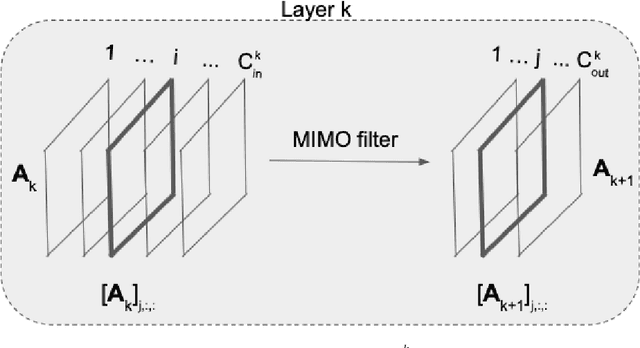
Abstract:Machine learning frameworks such as graph neural networks typically rely on a given, fixed graph to exploit relational inductive biases and thus effectively learn from network data. However, when said graphs are (partially) unobserved, noisy, or dynamic, the problem of inferring graph structure from data becomes relevant. In this paper, we postulate a graph convolutional relationship between the observed and latent graphs, and formulate the graph learning task as a network inverse (deconvolution) problem. In lieu of eigendecomposition-based spectral methods or iterative optimization solutions, we unroll and truncate proximal gradient iterations to arrive at a parameterized neural network architecture that we call a Graph Deconvolution Network (GDN). GDNs can learn a distribution of graphs in a supervised fashion, perform link prediction or edge-weight regression tasks by adapting the loss function, and they are inherently inductive. We corroborate GDN's superior graph recovery performance and its generalization to larger graphs using synthetic data in supervised settings. Furthermore, we demonstrate the robustness and representation power of GDNs on real world neuroimaging and social network datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge