Mason T. Chen
Rapid tissue oxygenation mapping from snapshot structured-light images with adversarial deep learning
Jul 01, 2020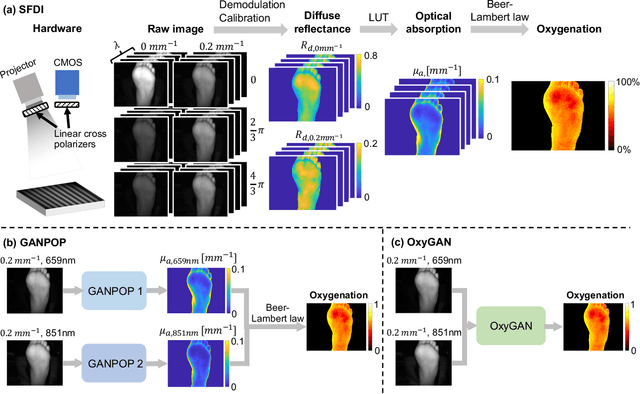
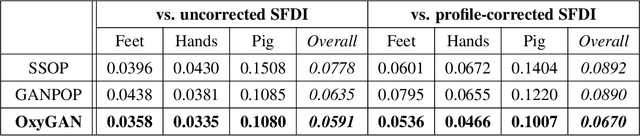
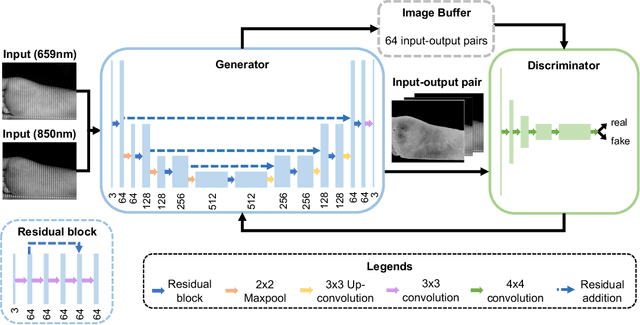
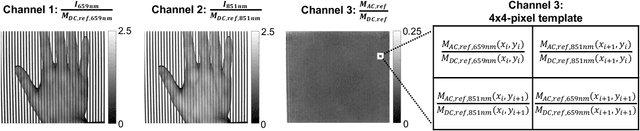
Abstract:Spatial frequency domain imaging (SFDI) is a powerful technique for mapping tissue oxygen saturation over a wide field of view. However, current SFDI methods either require a sequence of several images with different illumination patterns or, in the case of single snapshot optical properties (SSOP), introduce artifacts and sacrifice accuracy. To avoid this tradeoff, we introduce OxyGAN: a data-driven, content-aware method to estimate tissue oxygenation directly from single structured light images using end-to-end generative adversarial networks. Conventional SFDI is used to obtain ground truth tissue oxygenation maps for ex vivo human esophagi, in vivo hands and feet, and an in vivo pig colon sample under 659 nm and 851 nm sinusoidal illumination. We benchmark OxyGAN by comparing to SSOP and to a two-step hybrid technique that uses a previously-developed deep learning model to predict optical properties followed by a physical model to calculate tissue oxygenation. When tested on human feet, a cross-validated OxyGAN maps tissue oxygenation with an accuracy of 96.5%. When applied to sample types not included in the training set, such as human hands and pig colon, OxyGAN achieves a 93.0% accuracy, demonstrating robustness to various tissue types. On average, OxyGAN outperforms SSOP and a hybrid model in estimating tissue oxygenation by 24.9% and 24.7%, respectively. Lastly, we optimize OxyGAN inference so that oxygenation maps are computed ~10 times faster than previous work, enabling video-rate, 25Hz imaging. Due to its rapid acquisition and processing speed, OxyGAN has the potential to enable real-time, high-fidelity tissue oxygenation mapping that may be useful for many clinical applications.
GANPOP: Generative Adversarial Network Prediction of Optical Properties from Single Snapshot Wide-field Images
Jun 20, 2019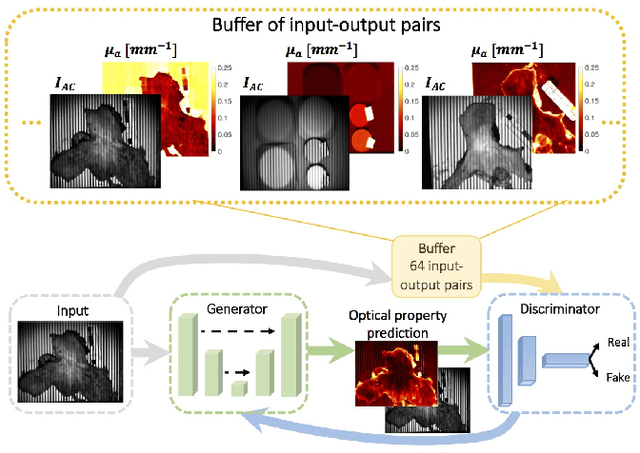
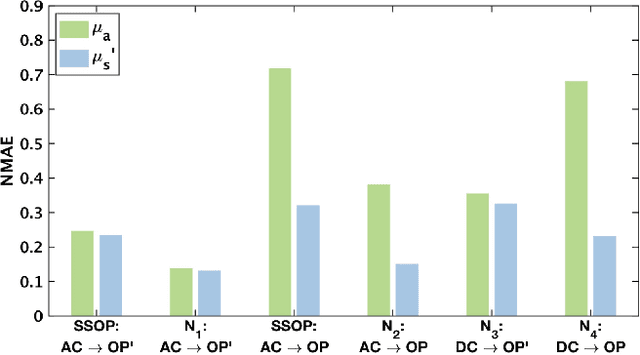
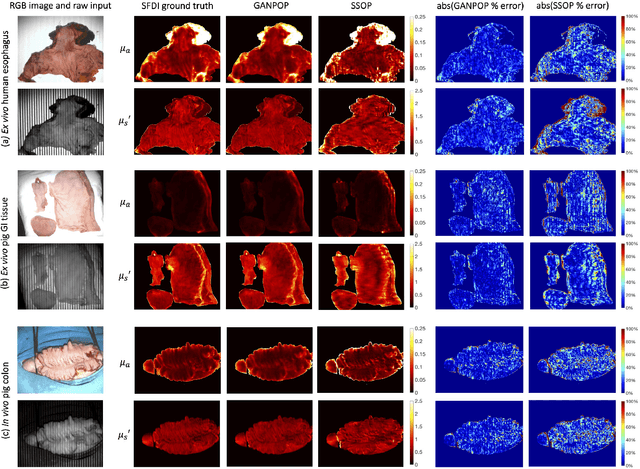
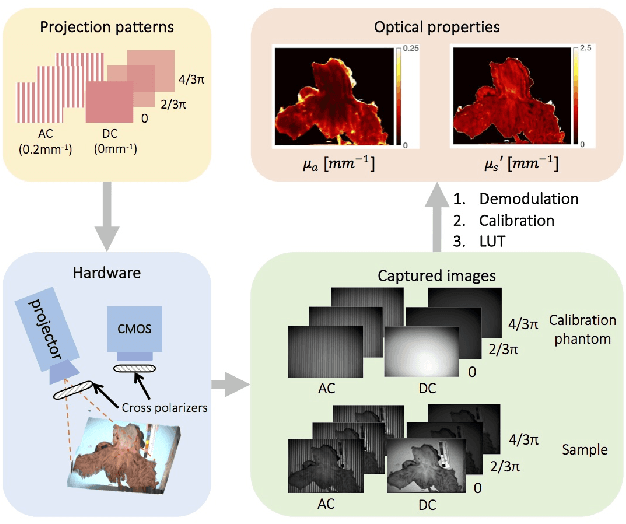
Abstract:We present a deep learning framework for wide-field, content-aware estimation of absorption and scattering coefficients of tissues, called Generative Adversarial Network Prediction of Optical Properties (GANPOP). Spatial frequency domain imaging is used to obtain ground-truth optical properties from in vivo human hands, freshly resected human esophagectomy samples and homogeneous tissue phantoms. Images of objects with either flat-field or structured illumination are paired with registered optical property maps and are used to train conditional generative adversarial networks that estimate optical properties from a single input image. We benchmark this approach by comparing GANPOP to a single-snapshot optical property (SSOP) technique, using a normalized mean absolute error (NMAE) metric. In human gastrointestinal specimens, GANPOP estimates both reduced scattering and absorption coefficients at 660 nm from a single 0.2/mm spatial frequency illumination image with 58% higher accuracy than SSOP. When applied to both in vivo and ex vivo swine tissues, a GANPOP model trained solely on human specimens and phantoms estimates optical properties with approximately 43% improvement over SSOP, indicating adaptability to sample variety. Moreover, we demonstrate that GANPOP estimates optical properties from flat-field illumination images with similar error to SSOP, which requires structured-illumination. Given a training set that appropriately spans the target domain, GANPOP has the potential to enable rapid and accurate wide-field measurements of optical properties, even from conventional imaging systems with flat-field illumination.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge