Mary Adewunmi
Enhancing Health Mention Classification Performance: A Study on Advancements in Parameter Efficient Tuning
Apr 30, 2025Abstract:Health Mention Classification (HMC) plays a critical role in leveraging social media posts for real-time tracking and public health monitoring. Nevertheless, the process of HMC presents significant challenges due to its intricate nature, primarily stemming from the contextual aspects of health mentions, such as figurative language and descriptive terminology, rather than explicitly reflecting a personal ailment. To address this problem, we argue that clearer mentions can be achieved through conventional fine-tuning with enhanced parameters of biomedical natural language methods (NLP). In this study, we explore different techniques such as the utilisation of part-of-speech (POS) tagger information, improving on PEFT techniques, and different combinations thereof. Extensive experiments are conducted on three widely used datasets: RHDM, PHM, and Illness. The results incorporated POS tagger information, and leveraging PEFT techniques significantly improves performance in terms of F1-score compared to state-of-the-art methods across all three datasets by utilising smaller models and efficient training. Furthermore, the findings highlight the effectiveness of incorporating POS tagger information and leveraging PEFT techniques for HMC. In conclusion, the proposed methodology presents a potentially effective approach to accurately classifying health mentions in social media posts while optimising the model size and training efficiency.
PubTrend: General Overview of Artificial Intelligence for Colorectal cancer diagnosis from 2010-2022
Jul 06, 2024


Abstract:Colorectal cancer (CRC) is among the most prevalent cancers in the world. Due to numerous scholarly papers and broad enquiries about specific use cases for artificial intelligence (AI) in colorectal cancer, researchers find it challenging to explore relevant papers on the current knowledge, comprehensive knowledge, and past methodologies in the literature review. This review extracts recent AI technology advances for diagnosing colorectal cancer from January 2010 to March 2022. PubTrends was used to identify and automate the intellectual structure and comparable papers on the use of AI in colorectal cancer diagnosis using the most cited papers, keywords, and similar papers. Papers with quantitative results were represented with a tabular summary, and other paper contributions were in a sentence summary. Twenty-four (24) out of the forty-nine (49) top-cited papers were quantitative results, with one (1) outlier about lung cancer comprehensive screening. The most frequently used words were: "polyps," "detected", "image," and "colonoscopy." In addition, 83 per cent of the terms frequently used shortly before 2022 were image, polyps, detected, colonoscopy, and learning. In addition, 16 per cent are preparation, variant, classification, sample, and surgery. The review showcases 49 of the 50 most cited papers, their notable contributions, objectives, specific AI methods, results, conclusions, and further recommendations. These papers highlight the limitations of colonoscopy for therapeutic use. The review concluded that despite the enormous benefits of using artificial intelligence, from improving diagnosis, the medical AI programmer still needs to be actively involved in the diagnosis team for effective results in CRC diagnosis.
Enhancing Transformer-Based Segmentation for Breast Cancer Diagnosis using Auto-Augmentation and Search Optimisation Techniques
Nov 18, 2023



Abstract:Breast cancer remains a critical global health challenge, necessitating early and accurate detection for effective treatment. This paper introduces a methodology that combines automated image augmentation selection (RandAugment) with search optimisation strategies (Tree-based Parzen Estimator) to identify optimal values for the number of image augmentations and the magnitude of their associated augmentation parameters, leading to enhanced segmentation performance. We empirically validate our approach on breast cancer histology slides, focusing on the segmentation of cancer cells. A comparative analysis of state-of-the-art transformer-based segmentation models is conducted, including SegFormer, PoolFormer, and MaskFormer models, to establish a comprehensive baseline, before applying the augmentation methodology. Our results show that the proposed methodology leads to segmentation models that are more resilient to variations in histology slides whilst maintaining high levels of segmentation performance, and show improved segmentation of the tumour class when compared to previous research. Our best result after applying the augmentations is a Dice Score of 84.08 and an IoU score of 72.54 when segmenting the tumour class. The primary contribution of this paper is the development of a methodology that enhances segmentation performance while ensuring model robustness to data variances. This has significant implications for medical practitioners, enabling the development of more effective machine learning models for clinical applications to identify breast cancer cells from histology slides. Furthermore, the codebase accompanying this research will be released upon publication. This will facilitate further research and application development based on our methodology, thereby amplifying its impact.
Improved Breast Cancer Diagnosis through Transfer Learning on Hematoxylin and Eosin Stained Histology Images
Sep 15, 2023

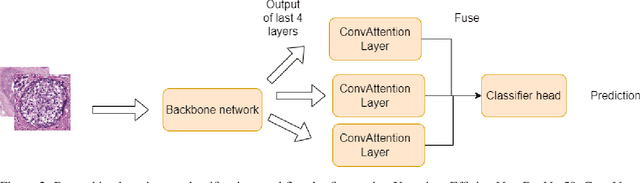
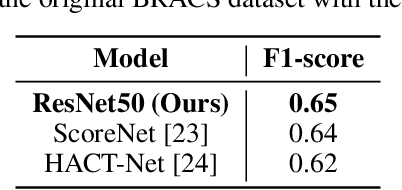
Abstract:Breast cancer is one of the leading causes of death for women worldwide. Early screening is essential for early identification, but the chance of survival declines as the cancer progresses into advanced stages. For this study, the most recent BRACS dataset of histological (H\&E) stained images was used to classify breast cancer tumours, which contains both the whole-slide images (WSI) and region-of-interest (ROI) images, however, for our study we have considered ROI images. We have experimented using different pre-trained deep learning models, such as Xception, EfficientNet, ResNet50, and InceptionResNet, pre-trained on the ImageNet weights. We pre-processed the BRACS ROI along with image augmentation, upsampling, and dataset split strategies. For the default dataset split, the best results were obtained by ResNet50 achieving 66\% f1-score. For the custom dataset split, the best results were obtained by performing upsampling and image augmentation which results in 96.2\% f1-score. Our second approach also reduced the number of false positive and false negative classifications to less than 3\% for each class. We believe that our study significantly impacts the early diagnosis and identification of breast cancer tumors and their subtypes, especially atypical and malignant tumors, thus improving patient outcomes and reducing patient mortality rates. Overall, this study has primarily focused on identifying seven (7) breast cancer tumor subtypes, and we believe that the experimental models can be fine-tuned further to generalize over previous breast cancer histology datasets as well.
Artificial intelligence based prediction on lung cancer risk factors using deep learning
Apr 11, 2023Abstract:In this proposed work, we identified the significant research issues on lung cancer risk factors. Capturing and defining symptoms at an early stage is one of the most difficult phases for patients. Based on the history of patients records, we reviewed a number of current research studies on lung cancer and its various stages. We identified that lung cancer is one of the significant research issues in predicting the early stages of cancer disease. This research aimed to develop a model that can detect lung cancer with a remarkably high level of accuracy using the deep learning approach (convolution neural network). This method considers and resolves significant gaps in previous studies. We compare the accuracy levels and loss values of our model with VGG16, InceptionV3, and Resnet50. We found that our model achieved an accuracy of 94% and a minimum loss of 0.1%. Hence physicians can use our convolution neural network models for predicting lung cancer risk factors in the real world. Moreover, this investigation reveals that squamous cell carcinoma, normal, adenocarcinoma, and large cell carcinoma are the most significant risk factors. In addition, the remaining attributes are also crucial for achieving the best performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge