Fahad Ahmed
From drift to adaptation to the failed ml model: Transfer Learning in Industrial MLOps
Feb 01, 2026Abstract:Model adaptation to production environment is critical for reliable Machine Learning Operations (MLOps), less attention is paid to developing systematic framework for updating the ML models when they fail under data drift. This paper compares the transfer learning enabled model update strategies including ensemble transfer learning (ETL), all-layers transfer learning (ALTL), and last-layer transfer learning (LLTL) for updating the failed feedforward artificial neural network (ANN) model. The flue gas differential pressure across the air preheater unit installed in a 660 MW thermal power plant is analyzed as a case study since it mimics the batch processes due to load cycling in the power plant. Updating the failed ANN model by three transfer learning techniques reveals that ETL provides relatively higher predictive accuracy for the batch size of 5 days than those of LLTL and ALTL. However, ALTL is found to be suitable for effective update of the model trained on large batch size (8 days). A mixed trend is observed for computational requirement (hyperparameter tuning and model training) of model update techniques for different batch sizes. These fundamental and empiric insights obtained from the batch process-based industrial case study can assist the MLOps practitioners in adapting the failed models to data drifts for the accurate monitoring of industrial processes.
Enhancing Transformer-Based Segmentation for Breast Cancer Diagnosis using Auto-Augmentation and Search Optimisation Techniques
Nov 18, 2023



Abstract:Breast cancer remains a critical global health challenge, necessitating early and accurate detection for effective treatment. This paper introduces a methodology that combines automated image augmentation selection (RandAugment) with search optimisation strategies (Tree-based Parzen Estimator) to identify optimal values for the number of image augmentations and the magnitude of their associated augmentation parameters, leading to enhanced segmentation performance. We empirically validate our approach on breast cancer histology slides, focusing on the segmentation of cancer cells. A comparative analysis of state-of-the-art transformer-based segmentation models is conducted, including SegFormer, PoolFormer, and MaskFormer models, to establish a comprehensive baseline, before applying the augmentation methodology. Our results show that the proposed methodology leads to segmentation models that are more resilient to variations in histology slides whilst maintaining high levels of segmentation performance, and show improved segmentation of the tumour class when compared to previous research. Our best result after applying the augmentations is a Dice Score of 84.08 and an IoU score of 72.54 when segmenting the tumour class. The primary contribution of this paper is the development of a methodology that enhances segmentation performance while ensuring model robustness to data variances. This has significant implications for medical practitioners, enabling the development of more effective machine learning models for clinical applications to identify breast cancer cells from histology slides. Furthermore, the codebase accompanying this research will be released upon publication. This will facilitate further research and application development based on our methodology, thereby amplifying its impact.
Improved Breast Cancer Diagnosis through Transfer Learning on Hematoxylin and Eosin Stained Histology Images
Sep 15, 2023

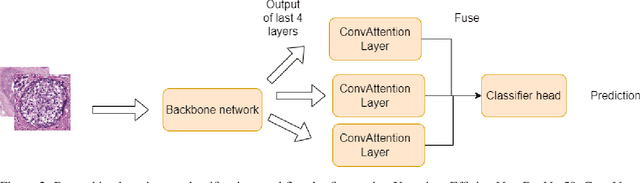
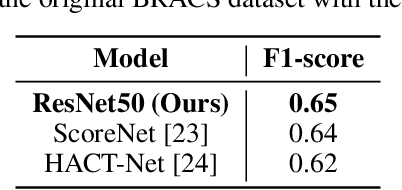
Abstract:Breast cancer is one of the leading causes of death for women worldwide. Early screening is essential for early identification, but the chance of survival declines as the cancer progresses into advanced stages. For this study, the most recent BRACS dataset of histological (H\&E) stained images was used to classify breast cancer tumours, which contains both the whole-slide images (WSI) and region-of-interest (ROI) images, however, for our study we have considered ROI images. We have experimented using different pre-trained deep learning models, such as Xception, EfficientNet, ResNet50, and InceptionResNet, pre-trained on the ImageNet weights. We pre-processed the BRACS ROI along with image augmentation, upsampling, and dataset split strategies. For the default dataset split, the best results were obtained by ResNet50 achieving 66\% f1-score. For the custom dataset split, the best results were obtained by performing upsampling and image augmentation which results in 96.2\% f1-score. Our second approach also reduced the number of false positive and false negative classifications to less than 3\% for each class. We believe that our study significantly impacts the early diagnosis and identification of breast cancer tumors and their subtypes, especially atypical and malignant tumors, thus improving patient outcomes and reducing patient mortality rates. Overall, this study has primarily focused on identifying seven (7) breast cancer tumor subtypes, and we believe that the experimental models can be fine-tuned further to generalize over previous breast cancer histology datasets as well.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge