Martin Strauch
Semi-supervised Instance Segmentation with a Learned Shape Prior
Sep 09, 2023Abstract:To date, most instance segmentation approaches are based on supervised learning that requires a considerable amount of annotated object contours as training ground truth. Here, we propose a framework that searches for the target object based on a shape prior. The shape prior model is learned with a variational autoencoder that requires only a very limited amount of training data: In our experiments, a few dozens of object shape patches from the target dataset, as well as purely synthetic shapes, were sufficient to achieve results en par with supervised methods with full access to training data on two out of three cell segmentation datasets. Our method with a synthetic shape prior was superior to pre-trained supervised models with access to limited domain-specific training data on all three datasets. Since the learning of prior models requires shape patches, whether real or synthetic data, we call this framework semi-supervised learning.
High-throughput Phenotyping of Nematode Cysts
Oct 13, 2021
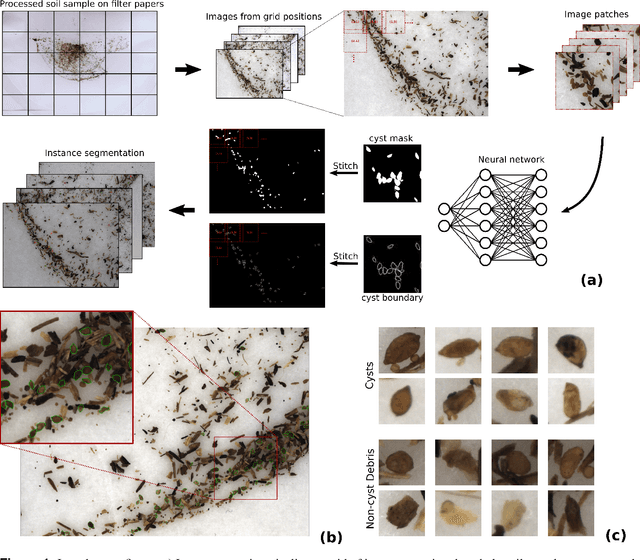
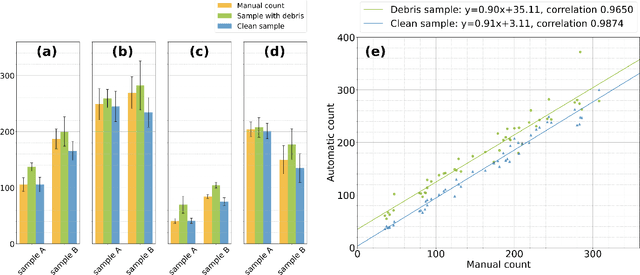

Abstract:The beet cyst nematode (BCN) Heterodera schachtii is a plant pest responsible for crop loss on a global scale. Here, we introduce a high-throughput system based on computer vision that allows quantifying BCN infestation and characterizing nematode cysts through phenotyping. After recording microscopic images of soil extracts in a standardized setting, an instance segmentation algorithm serves to detect nematode cysts in these samples. Going beyond fast and precise cyst counting, the image-based approach enables quantification of cyst density and phenotyping of morphological features of cysts under different conditions, providing the basis for high-throughput applications in agriculture and plant breeding research.
Instance Segmentation of Biomedical Images with an Object-aware Embedding Learned with Local Constraints
Apr 21, 2020



Abstract:Automatic instance segmentation is a problem that occurs in many biomedical applications. State-of-the-art approaches either perform semantic segmentation or refine object bounding boxes obtained from detection methods. Both suffer from crowded objects to varying degrees, merging adjacent objects or suppressing a valid object. In this work, we assign an embedding vector to each pixel through a deep neural network. The network is trained to output embedding vectors of similar directions for pixels from the same object, while adjacent objects are orthogonal in the embedding space, which effectively avoids the fusion of objects in a crowd. Our method yields state-of-the-art results even with a light-weighted backbone network on a cell segmentation (BBBC006 + DSB2018) and a leaf segmentation data set (CVPPP2017). The code and model weights are public available.
* MICCAI 2019
A CNN Framenwork Based on Line Annotations for Detecting Nematodes in Microscopic Images
Apr 21, 2020


Abstract:Plant parasitic nematodes cause damage to crop plants on a global scale. Robust detection on image data is a prerequisite for monitoring such nematodes, as well as for many biological studies involving the nematode C. elegans, a common model organism. Here, we propose a framework for detecting worm-shaped objects in microscopic images that is based on convolutional neural networks (CNNs). We annotate nematodes with curved lines along the body, which is more suitable for worm-shaped objects than bounding boxes. The trained model predicts worm skeletons and body endpoints. The endpoints serve to untangle the skeletons from which segmentation masks are reconstructed by estimating the body width at each location along the skeleton. With light-weight backbone networks, we achieve 75.85 % precision, 73.02 % recall on a potato cyst nematode data set and 84.20 % precision, 85.63 % recall on a public C. elegans data set.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge