Martin Kang
Quantification of BERT Diagnosis Generalizability Across Medical Specialties Using Semantic Dataset Distance
Aug 20, 2020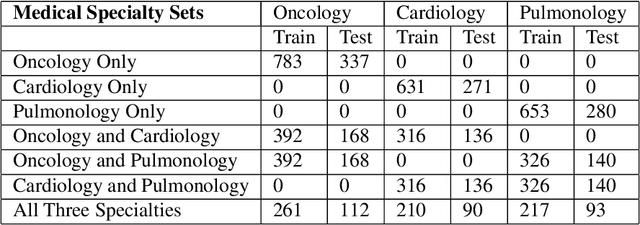


Abstract:Deep learning models in healthcare may fail to generalize on data from unseen corpora. Additionally, no quantitative metric exists to tell how existing models will perform on new data. Previous studies demonstrated that NLP models of medical notes generalize variably between institutions, but ignored other levels of healthcare organization. We measured SciBERT diagnosis sentiment classifier generalizability between medical specialties using EHR sentences from MIMIC-III. Models trained on one specialty performed better on internal test sets than mixed or external test sets (mean AUCs 0.92, 0.87, and 0.83, respectively; p = 0.016). When models are trained on more specialties, they have better test performances (p < 1e-4). Model performance on new corpora is directly correlated to the similarity between train and test sentence content (p < 1e-4). Future studies should assess additional axes of generalization to ensure deep learning models fulfil their intended purpose across institutions, specialties, and practices.
Augmented Curation of Unstructured Clinical Notes from a Massive EHR System Reveals Specific Phenotypic Signature of Impending COVID-19 Diagnosis
Apr 28, 2020

Abstract:Understanding the temporal dynamics of COVID-19 patient phenotypes is necessary to derive fine-grained resolution of pathophysiology. Here we use state-of-the-art deep neural networks over an institution-wide machine intelligence platform for the augmented curation of 15.8 million clinical notes from 30,494 patients subjected to COVID-19 PCR diagnostic testing. By contrasting the Electronic Health Record (EHR)-derived clinical phenotypes of COVID-19-positive (COVIDpos, n=635) versus COVID-19-negative (COVIDneg, n=29,859) patients over each day of the week preceding the PCR testing date, we identify anosmia/dysgeusia (37.4-fold), myalgia/arthralgia (2.6-fold), diarrhea (2.2-fold), fever/chills (2.1-fold), respiratory difficulty (1.9-fold), and cough (1.8-fold) as significantly amplified in COVIDpos over COVIDneg patients. The specific combination of cough and diarrhea has a 3.2-fold amplification in COVIDpos patients during the week prior to PCR testing, and along with anosmia/dysgeusia, constitutes the earliest EHR-derived signature of COVID-19 (4-7 days prior to typical PCR testing date). This study introduces an Augmented Intelligence platform for the real-time synthesis of institutional knowledge captured in EHRs. The platform holds tremendous potential for scaling up curation throughput, with minimal need for retraining underlying neural networks, thus promising EHR-powered early diagnosis for a broad spectrum of diseases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge