Martin A. Lindquist
Department of Biostatistics, Johns Hopkins University, USA
Mode decomposition-based time-varying phase synchronization for fMRI Data
Mar 26, 2022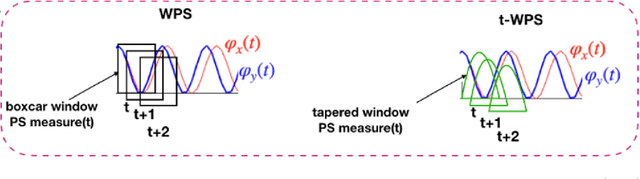
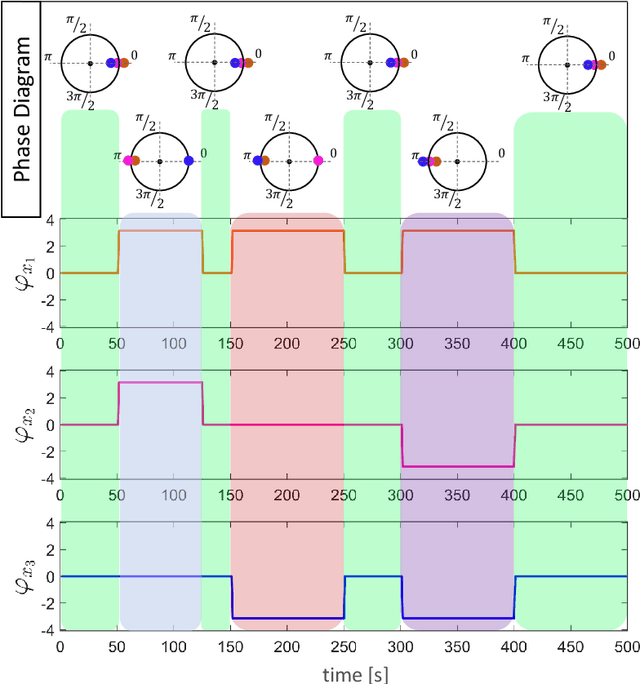
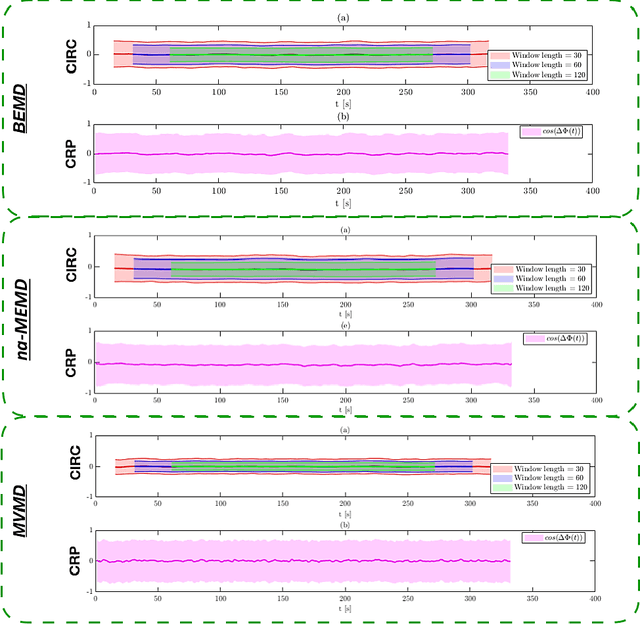
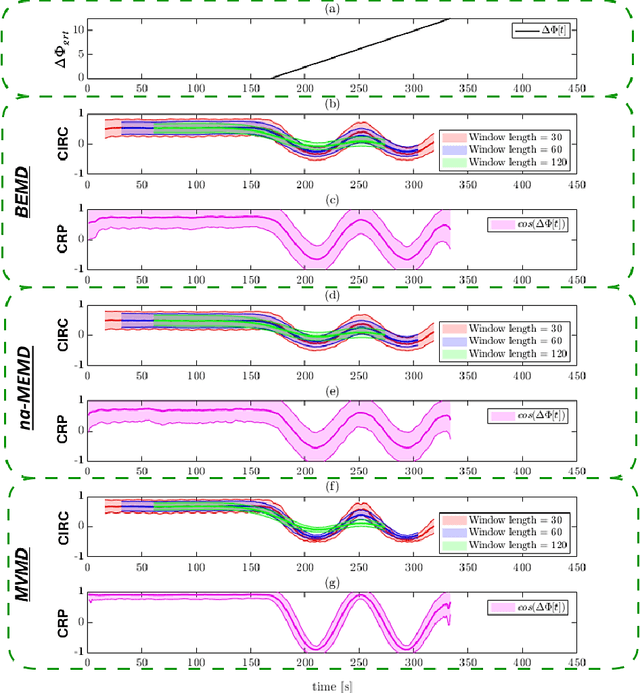
Abstract:Recently there has been significant interest in measuring time-varying functional connectivity (TVC) between different brain regions using resting-state functional magnetic resonance imaging (rs-fMRI) data. One way to assess the relationship between signals from different brain regions is to measure their phase synchronization (PS) across time. However, this requires the \textit{a priori} choice of type and cut-off frequencies for the bandpass filter needed to perform the analysis. Here we explore alternative approaches based on the use of various mode decomposition (MD) techniques that circumvent this issue. These techniques allow for the data driven decomposition of signals jointly into narrow-band components at different frequencies, thus fulfilling the requirements needed to measure PS. We explore several variants of MD, including empirical mode decomposition (EMD), bivariate EMD (BEMD), noise-assisted multivariate EMD (na-MEMD), and introduce the use of multivariate variational mode decomposition (MVMD) in the context of estimating time-varying PS. We contrast the approaches using a series of simulations and application to rs-fMRI data. Our results show that MVMD outperforms other evaluated MD approaches, and further suggests that this approach can be used as a tool to reliably investigate time-varying PS in rs-fMRI data.
Evaluating phase synchronization methods in fMRI: a comparison study and new approaches
Sep 21, 2020



Abstract:In recent years there has been growing interest in measuring time-varying functional connectivity between different brain regions using resting-state functional magnetic resonance imaging (rs-fMRI) data. One way to assess the relationship between signals from different brain regions is to measure their phase synchronization (PS) across time. There are several ways to perform such analyses, and here we compare methods that utilize a PS metric together with a sliding window, referred to here as windowed phase synchronization (WPS), with those that directly measure the instantaneous phase synchronization (IPS). In particular, IPS has recently gained popularity as it offers single time-point resolution of time-resolved fMRI connectivity. In this paper, we discuss the underlying assumptions required for performing PS analyses and emphasize the necessity of band-pass filtering the data to obtain valid results. We review various methods for evaluating PS and introduce a new approach within the IPS framework denoted the cosine of the relative phase (CRP). We contrast methods through a series of simulations and application to rs-fMRI data. Our results indicate that CRP outperforms other tested methods and overcomes issues related to undetected temporal transitions from positive to negative associations common in IPS analysis. Further, in contrast to phase coherence, CRP unfolds the distribution of PS measures, which benefits subsequent clustering of PS matrices into recurring brain states.
Automatic Identification of Twin Zygosity in Resting-State Functional MRI
Oct 26, 2018



Abstract:A key strength of twin studies arises from the fact that there are two types of twins, monozygotic and dizygotic, that share differing amounts of genetic information. Accurate differentiation of twin types allows efficient inference on genetic influences in a population. However, identification of zygosity is often prone to errors without genotying. In this study, we propose a novel pairwise feature representation to classify the zygosity of twin pairs of resting state functional magnetic resonance images (rs-fMRI). For this, we project an fMRI signal to a set of basis functions and use the projection coefficients as the compact and discriminative feature representation of noisy fMRI. We encode the relationship between twins as the correlation between the new feature representations across brain regions. We employ hill climbing variable selection to identify brain regions that are the most genetically affected. The proposed framework was applied to 208 twin pairs and achieved 94.19% classification accuracy in automatically identifying the zygosity of paired images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge