Marie Beurton-Aimar
Multitask Multi-database Emotion Recognition
Jul 12, 2021



Abstract:In this work, we introduce our submission to the 2nd Affective Behavior Analysis in-the-wild (ABAW) 2021 competition. We train a unified deep learning model on multi-databases to perform two tasks: seven basic facial expressions prediction and valence-arousal estimation. Since these databases do not contains labels for all the two tasks, we have applied the distillation knowledge technique to train two networks: one teacher and one student model. The student model will be trained using both ground truth labels and soft labels derived from the pretrained teacher model. During the training, we add one more task, which is the combination of the two mentioned tasks, for better exploiting inter-task correlations. We also exploit the sharing videos between the two tasks of the AffWild2 database that is used in the competition, to further improve the performance of the network. Experiment results shows that the network have achieved promising results on the validation set of the AffWild2 database. Code and pretrained model are publicly available at https://github.com/glmanhtu/multitask-abaw-2021
Using Graph Neural Networks to Reconstruct Ancient Documents
Nov 13, 2020



Abstract:In recent years, machine learning and deep learning approaches such as artificial neural networks have gained in popularity for the resolution of automatic puzzle resolution problems. Indeed, these methods are able to extract high-level representations from images, and then can be trained to separate matching image pieces from non-matching ones. These applications have many similarities to the problem of ancient document reconstruction from partially recovered fragments. In this work we present a solution based on a Graph Neural Network, using pairwise patch information to assign labels to edges representing the spatial relationships between pairs. This network classifies the relationship between a source and a target patch as being one of Up, Down, Left, Right or None. By doing so for all edges, our model outputs a new graph representing a reconstruction proposal. Finally, we show that our model is not only able to provide correct classifications at the edge-level, but also to generate partial or full reconstruction graphs from a set of patches.
Predicting Brain Degeneration with a Multimodal Siamese Neural Network
Nov 02, 2020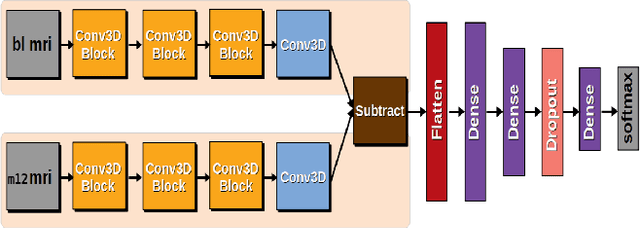
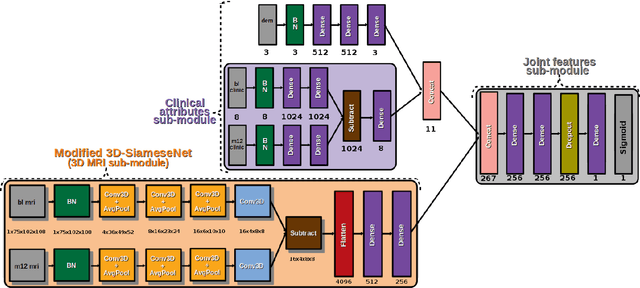
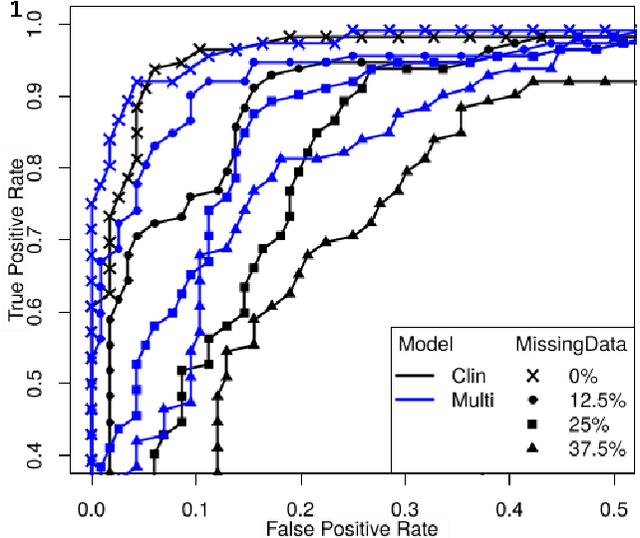
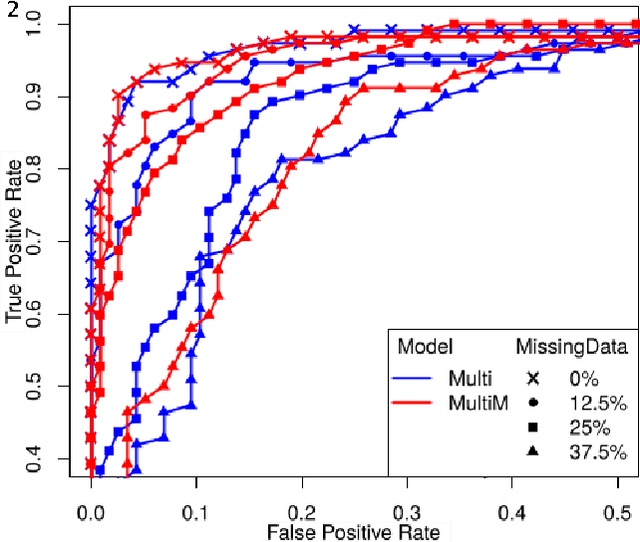
Abstract:To study neurodegenerative diseases, longitudinal studies are carried on volunteer patients. During a time span of several months to several years, they go through regular medical visits to acquire data from different modalities, such as biological samples, cognitive tests, structural and functional imaging. These variables are heterogeneous but they all depend on the patient's health condition, meaning that there are possibly unknown relationships between all modalities. Some information may be specific to some modalities, others may be complementary, and others may be redundant. Some data may also be missing. In this work we present a neural network architecture for multimodal learning, able to use imaging and clinical data from two time points to predict the evolution of a neurodegenerative disease, and robust to missing values. Our multimodal network achieves 92.5\% accuracy and an AUC score of 0.978 over a test set of 57 subjects. We also show the superiority of the multimodal architecture, for up to 37.5\% of missing values in test set subjects' clinical measurements, compared to a model using only the clinical modality.
3DSiameseNet to Analyze Brain MRI
Sep 03, 2019



Abstract:Prediction of the cognitive evolution of a person susceptible to develop a neurodegenerative disorder is crucial to provide an appropriate treatment as soon as possible. In this paper we propose a 3D siamese network designed to extract features from whole-brain 3D MRI images. We show that it is possible to extract meaningful features using convolution layers, reducing the need of classical image processing operations such as segmentation or pre-computing features such as cortical thickness. To lead this study we used the Alzheimer's Disease Neuroimaging Initiative (ADNI), a public data base of 3D MRI brain images. A set of 247 subjects has been extracted, all of the subjects having 2 images in a range of 12 months. In order to measure the evolution of the patients states we have compared these 2 images. Our work has been inspired at the beginning by an article of Bhagwat et al. in 2018, who have proposed a siamese network to predict the status of patients but without any convolutional layers and reducing the MRI images to a vector of features extracted from predefined ROIs. We show that our network achieves an accuracy of 90\% in the classification of cognitively declining VS stable patients. This result has been obtained without the help of a cognitive score and with a small number of patients comparing to the current datasets size claimed in deep learning domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge