Marianne Johansson Jørgensen
The Consequences of the Framing of Machine Learning Risk Prediction Models: Evaluation of Sepsis in General Wards
Jan 26, 2021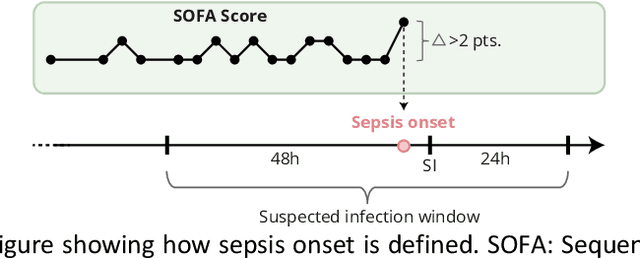

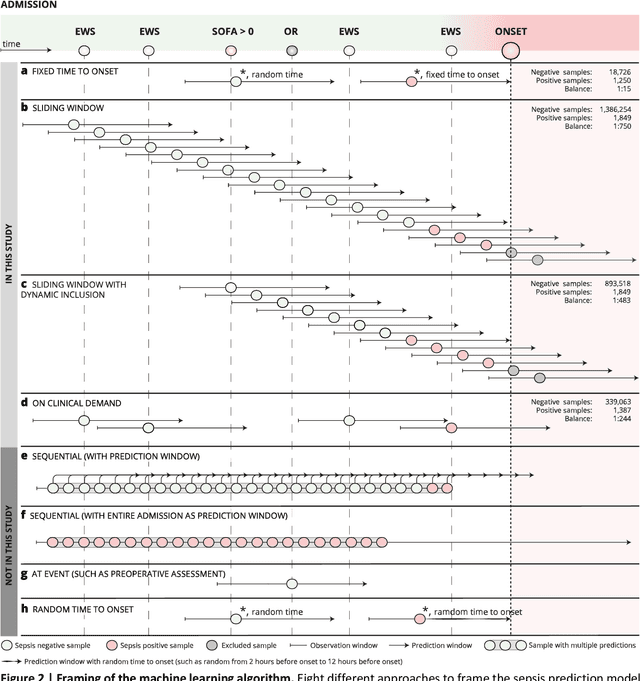
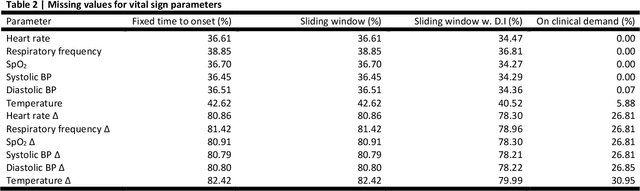
Abstract:Objectives: To evaluate the consequences of the framing of machine learning risk prediction models. We evaluate how framing affects model performance and model learning in four different approaches previously applied in published artificial-intelligence (AI) models. Setting and participants: We analysed structured secondary healthcare data from 221,283 citizens from four Danish municipalities who were 18 years of age or older. Results: The four models had similar population level performance (a mean area under the receiver operating characteristic curve of 0.73 to 0.82), in contrast to the mean average precision, which varied greatly from 0.007 to 0.385. Correspondingly, the percentage of missing values also varied between framing approaches. The on-clinical-demand framing, which involved samples for each time the clinicians made an early warning score assessment, showed the lowest percentage of missing values among the vital sign parameters, and this model was also able to learn more temporal dependencies than the others. The Shapley additive explanations demonstrated opposing interpretations of SpO2 in the prediction of sepsis as a consequence of differentially framed models. Conclusions: The profound consequences of framing mandate attention from clinicians and AI developers, as the understanding and reporting of framing are pivotal to the successful development and clinical implementation of future AI technology. Model framing must reflect the expected clinical environment. The importance of proper problem framing is by no means exclusive to sepsis prediction and applies to most clinical risk prediction models.
Explainable artificial intelligence model to predict acute critical illness from electronic health records
Dec 03, 2019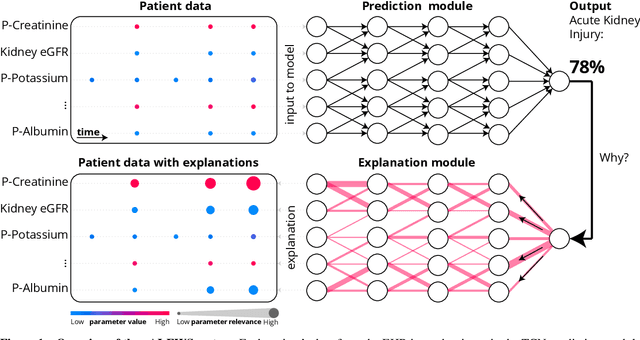
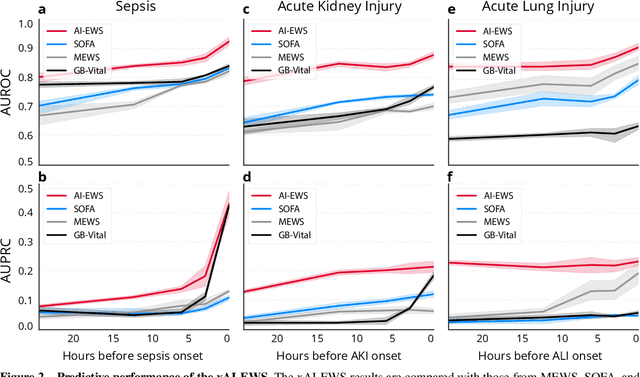
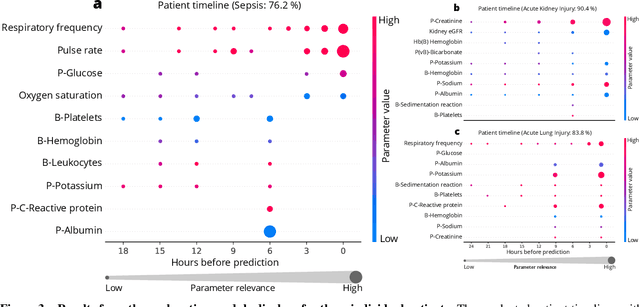
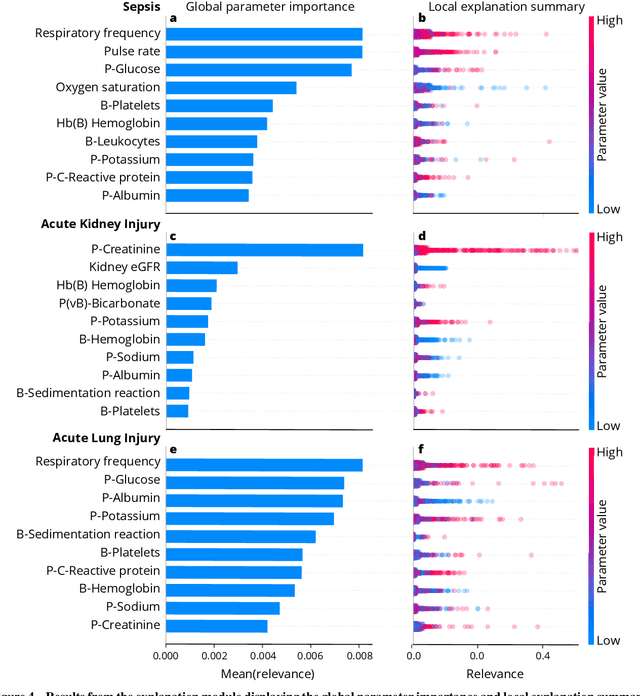
Abstract:We developed an explainable artificial intelligence (AI) early warning score (xAI-EWS) system for early detection of acute critical illness. While maintaining a high predictive performance, our system explains to the clinician on which relevant electronic health records (EHRs) data the prediction is grounded. Acute critical illness is often preceded by deterioration of routinely measured clinical parameters, e.g., blood pressure and heart rate. Early clinical prediction is typically based on manually calculated screening metrics that simply weigh these parameters, such as Early Warning Scores (EWS). The predictive performance of EWSs yields a tradeoff between sensitivity and specificity that can lead to negative outcomes for the patient. Previous work on EHR-trained AI systems offers promising results with high levels of predictive performance in relation to the early, real-time prediction of acute critical illness. However, without insight into the complex decisions by such system, clinical translation is hindered. In this letter, we present our xAI-EWS system, which potentiates clinical translation by accompanying a prediction with information on the EHR data explaining it.
Early detection of sepsis utilizing deep learning on electronic health record event sequences
Jun 07, 2019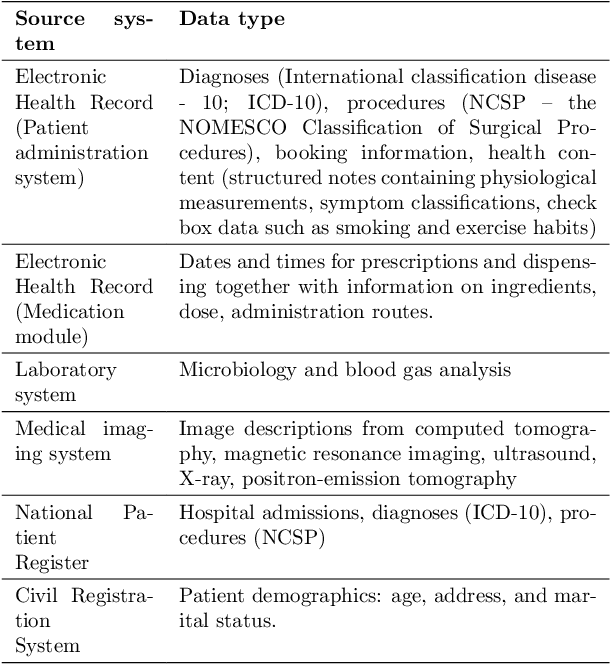
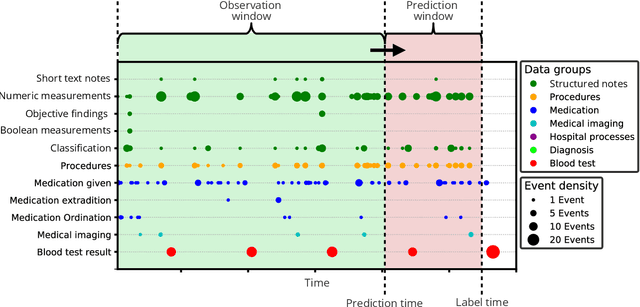
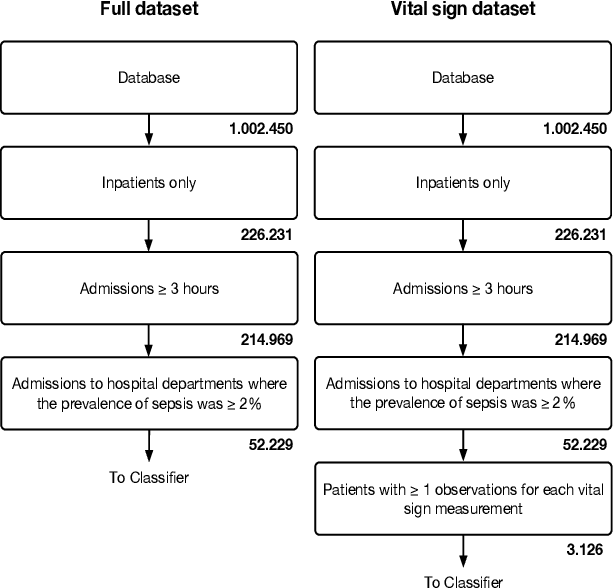
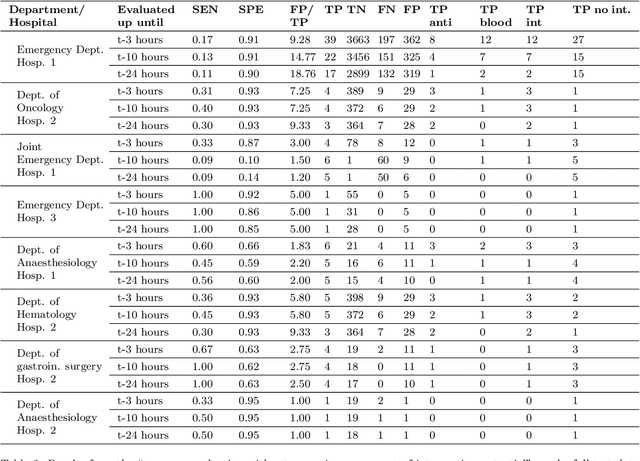
Abstract:The timeliness of detection of a sepsis event in progress is a crucial factor in the outcome for the patient. Machine learning models built from data in electronic health records can be used as an effective tool for improving this timeliness, but so far the potential for clinical implementations has been largely limited to studies in intensive care units. This study will employ a richer data set that will expand the applicability of these models beyond intensive care units. Furthermore, we will circumvent several important limitations that have been found in the literature: 1) Models are evaluated shortly before sepsis onset without considering interventions already initiated. 2) Machine learning models are built on a restricted set of clinical parameters, which are not necessarily measured in all departments. 3) Model performance is limited by current knowledge of sepsis, as feature interactions and time dependencies are hardcoded into the model. In this study, we present a model to overcome these shortcomings using a deep learning approach on a diverse multicenter data set. We used retrospective data from multiple Danish hospitals over a seven-year period. Our sepsis detection system is constructed as a combination of a convolutional neural network and a long short-term memory network. We suggest a retrospective assessment of interventions by looking at intravenous antibiotics and blood cultures preceding the prediction time. Results show performance ranging from AUROC 0.856 (3 hours before sepsis onset) to AUROC 0.756 (24 hours before sepsis onset). We present a deep learning system for early detection of sepsis that is able to learn characteristics of the key factors and interactions from the raw event sequence data itself, without relying on a labor-intensive feature extraction work.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge