Maria Altbach
Learning to segment with limited annotations: Self-supervised pretraining with regression and contrastive loss in MRI
May 26, 2022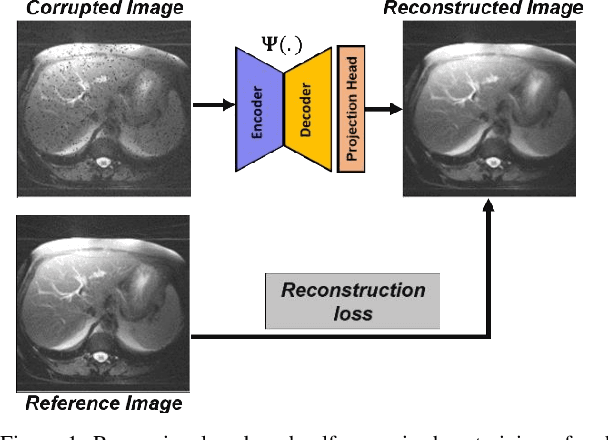
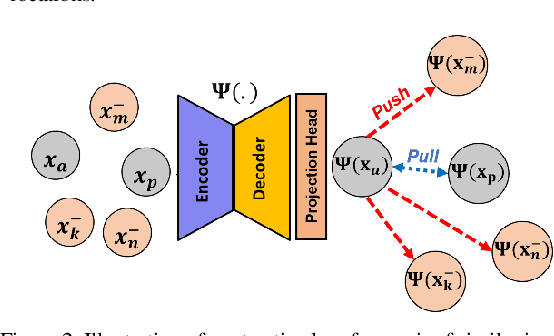
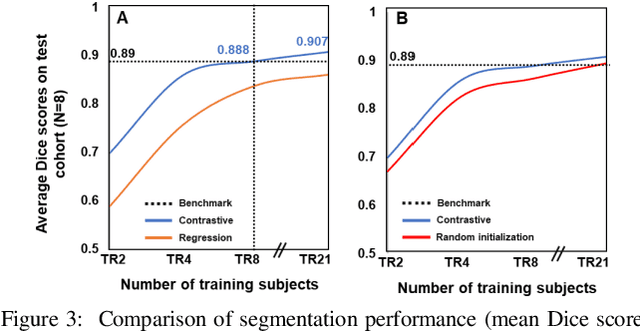
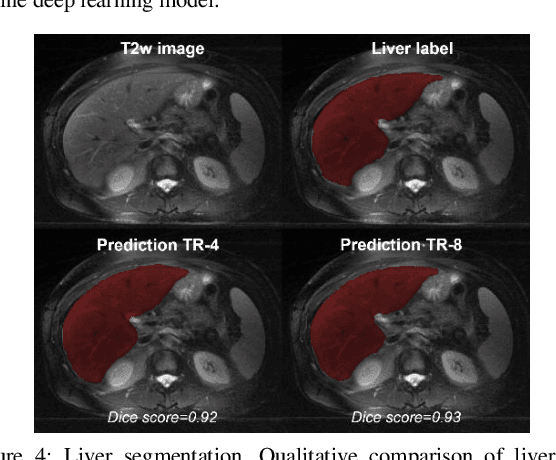
Abstract:Obtaining manual annotations for large datasets for supervised training of deep learning (DL) models is challenging. The availability of large unlabeled datasets compared to labeled ones motivate the use of self-supervised pretraining to initialize DL models for subsequent segmentation tasks. In this work, we consider two pre-training approaches for driving a DL model to learn different representations using: a) regression loss that exploits spatial dependencies within an image and b) contrastive loss that exploits semantic similarity between pairs of images. The effect of pretraining techniques is evaluated in two downstream segmentation applications using Magnetic Resonance (MR) images: a) liver segmentation in abdominal T2-weighted MR images and b) prostate segmentation in T2-weighted MR images of the prostate. We observed that DL models pretrained using self-supervision can be finetuned for comparable performance with fewer labeled datasets. Additionally, we also observed that initializing the DL model using contrastive loss based pretraining performed better than the regression loss.
A Cascaded Residual UNET for Fully Automated Segmentation of Prostate and Peripheral Zone in T2-weighted 3D Fast Spin Echo Images
Dec 25, 2020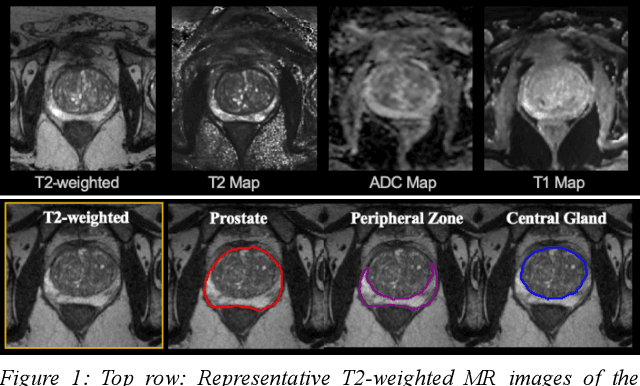
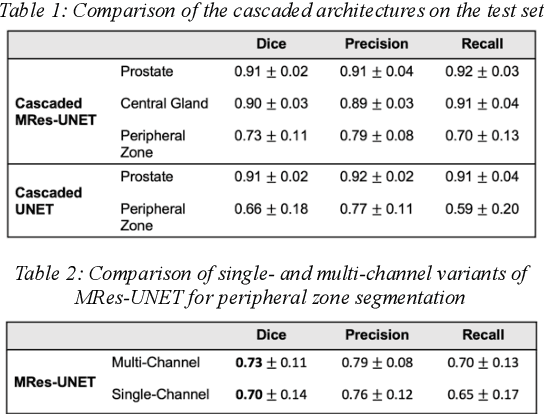
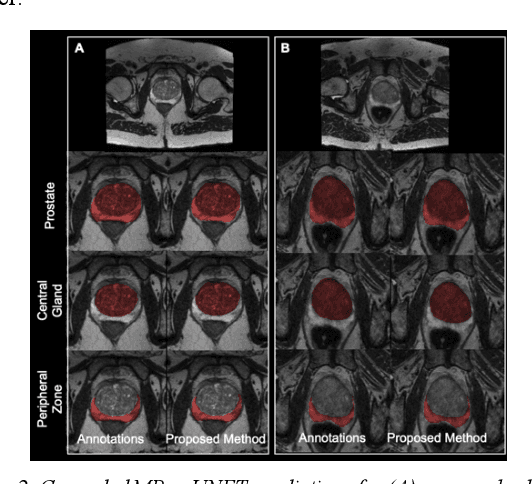
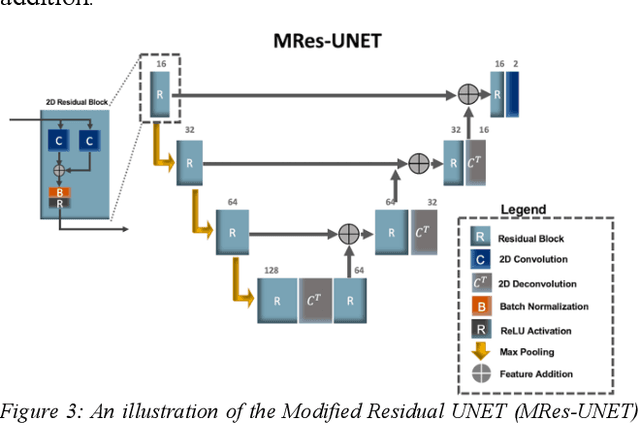
Abstract:Multi-parametric MR images have been shown to be effective in the non-invasive diagnosis of prostate cancer. Automated segmentation of the prostate eliminates the need for manual annotation by a radiologist which is time consuming. This improves efficiency in the extraction of imaging features for the characterization of prostate tissues. In this work, we propose a fully automated cascaded deep learning architecture with residual blocks, Cascaded MRes-UNET, for segmentation of the prostate gland and the peripheral zone in one pass through the network. The network yields high Dice scores ($0.91\pm.02$), precision ($0.91\pm.04$), and recall scores ($0.92\pm.03$) in prostate segmentation compared to manual annotations by an experienced radiologist. The average difference in total prostate volume estimation is less than 5%.
White matter hyperintensities volume and cognition: Assessment of a deep learning based lesion detection and quantification algorithm on the Alzheimers Disease Neuroimaging Initiative
Dec 24, 2020



Abstract:The relationship between cognition and white matter hyperintensities (WMH) volumes often depends on the accuracy of the lesion segmentation algorithm used. As such, accurate detection and quantification of WMH is of great interest. Here, we use a deep learning-based WMH segmentation algorithm, StackGen-Net, to detect and quantify WMH on 3D FLAIR volumes from ADNI. We used a subset of subjects (n=20) and obtained manual WMH segmentations by an experienced neuro-radiologist to demonstrate the accuracy of our algorithm. On a larger cohort of subjects (n=290), we observed that larger WMH volumes correlated with worse performance on executive function (P=.004), memory (P=.01), and language (P=.005).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge