Marcus Völp
I-GWAS: Privacy-Preserving Interdependent Genome-Wide Association Studies
Aug 17, 2022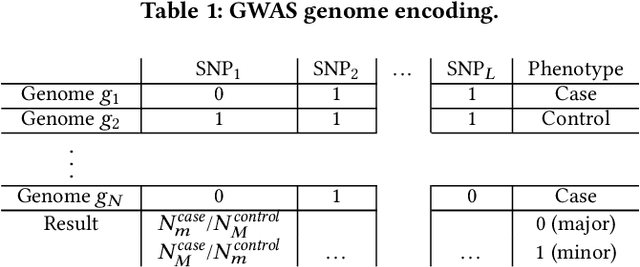
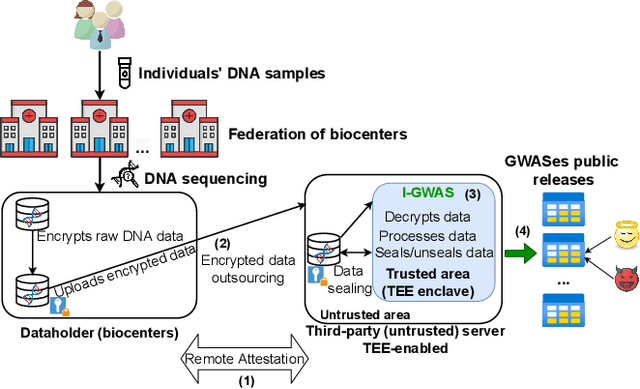
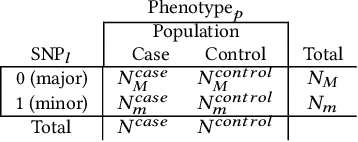
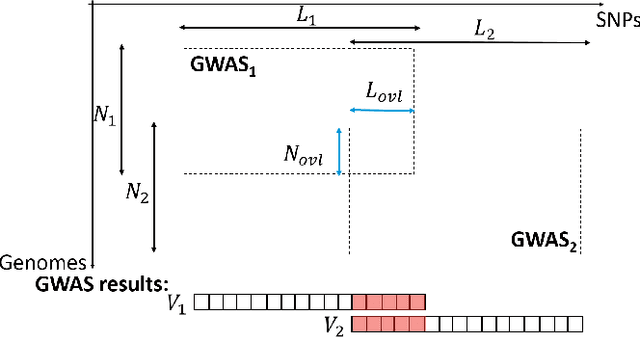
Abstract:Genome-wide Association Studies (GWASes) identify genomic variations that are statistically associated with a trait, such as a disease, in a group of individuals. Unfortunately, careless sharing of GWAS statistics might give rise to privacy attacks. Several works attempted to reconcile secure processing with privacy-preserving releases of GWASes. However, we highlight that these approaches remain vulnerable if GWASes utilize overlapping sets of individuals and genomic variations. In such conditions, we show that even when relying on state-of-the-art techniques for protecting releases, an adversary could reconstruct the genomic variations of up to 28.6% of participants, and that the released statistics of up to 92.3% of the genomic variations would enable membership inference attacks. We introduce I-GWAS, a novel framework that securely computes and releases the results of multiple possibly interdependent GWASes. I-GWAScontinuously releases privacy-preserving and noise-free GWAS results as new genomes become available.
Federated Geometric Monte Carlo Clustering to Counter Non-IID Datasets
Apr 23, 2022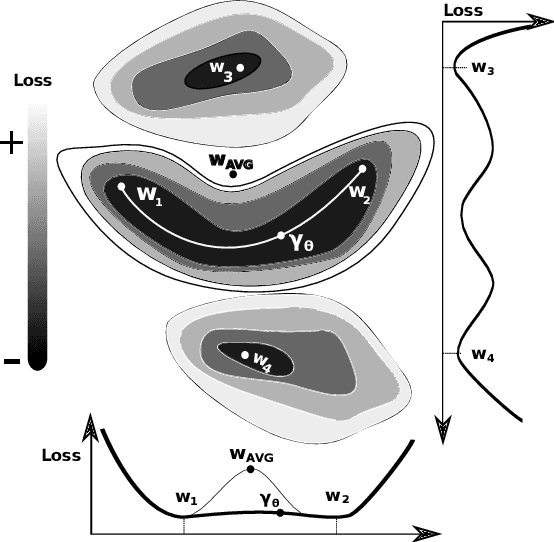


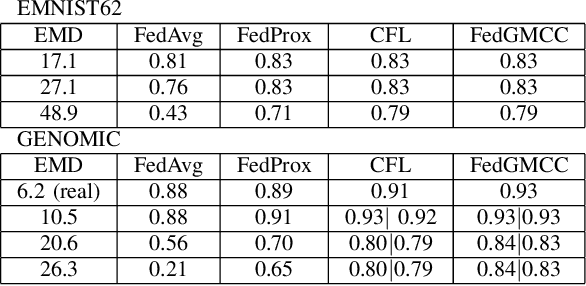
Abstract:Federated learning allows clients to collaboratively train models on datasets that are acquired in different locations and that cannot be exchanged because of their size or regulations. Such collected data is increasingly non-independent and non-identically distributed (non-IID), negatively affecting training accuracy. Previous works tried to mitigate the effects of non-IID datasets on training accuracy, focusing mainly on non-IID labels, however practical datasets often also contain non-IID features. To address both non-IID labels and features, we propose FedGMCC, a novel framework where a central server aggregates client models that it can cluster together. FedGMCC clustering relies on a Monte Carlo procedure that samples the output space of client models, infers their position in the weight space on a loss manifold and computes their geometric connection via an affine curve parametrization. FedGMCC aggregates connected models along their path connectivity to produce a richer global model, incorporating knowledge of all connected client models. FedGMCC outperforms FedAvg and FedProx in terms of convergence rates on the EMNIST62 and a genomic sequence classification datasets (by up to +63%). FedGMCC yields an improved accuracy (+4%) on the genomic dataset with respect to CFL, in high non-IID feature space settings and label incongruency.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge