Manyuan Lu
Anchoring to Exemplars for Training Mixture-of-Expert Cell Embeddings
Dec 06, 2021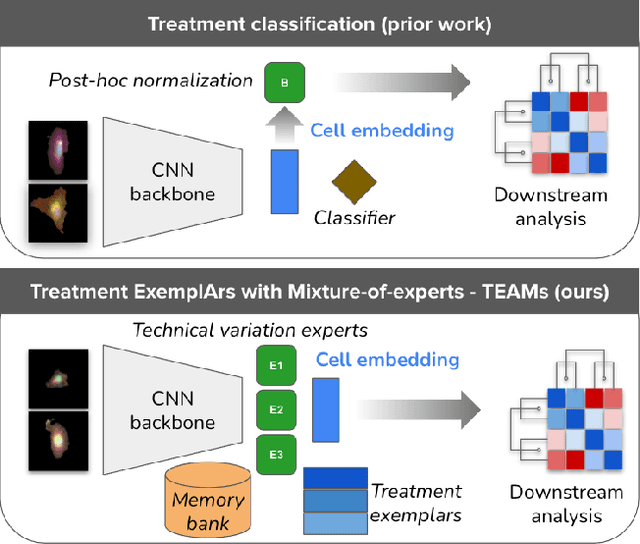
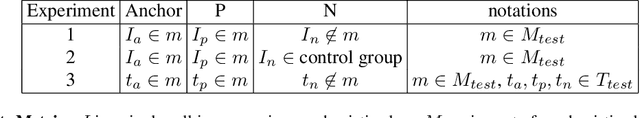
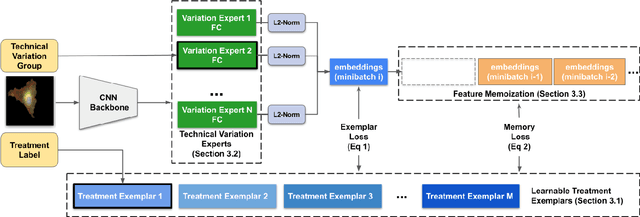
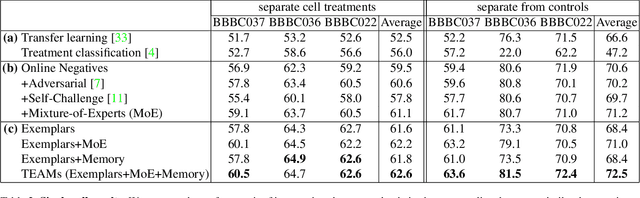
Abstract:Analyzing the morphology of cells in microscopy images can provide insights into the mechanism of compounds or the function of genes. Addressing this task requires methods that can not only extract biological information from the images, but also ignore technical variations, ie, changes in experimental procedure or differences between equipments used to collect microscopy images. We propose Treatment ExemplArs with Mixture-of-experts (TEAMs), an embedding learning approach that learns a set of experts that are specialized in capturing technical variations in our training set and then aggregates specialist's predictions at test time. Thus, TEAMs can learn powerful embeddings with less technical variation bias by minimizing the noise from every expert. To train our model, we leverage Treatment Exemplars that enable our approach to capture the distribution of the entire dataset in every minibatch while still fitting into GPU memory. We evaluate our approach on three datasets for tasks like drug discovery, boosting performance on identifying the true mechanism of action of cell treatments by 5.5-11% over the state-of-the-art.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge