Manu Vatish
PatchCTG: Patch Cardiotocography Transformer for Antepartum Fetal Health Monitoring
Nov 12, 2024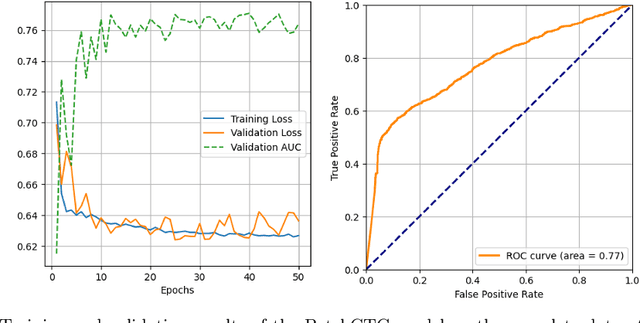
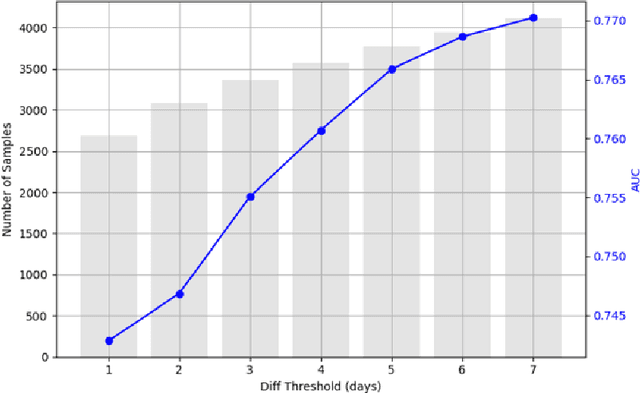
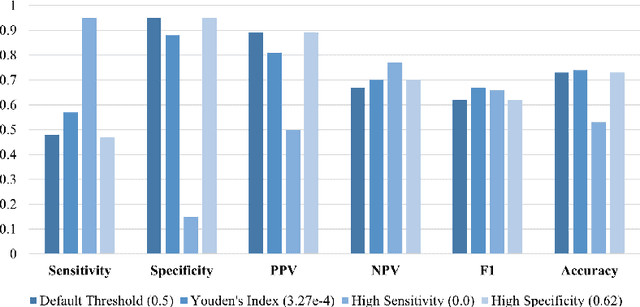
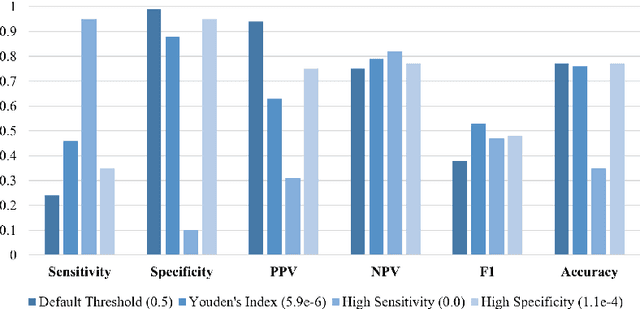
Abstract:Antepartum Cardiotocography (CTG) is vital for fetal health monitoring, but traditional methods like the Dawes-Redman system are often limited by high inter-observer variability, leading to inconsistent interpretations and potential misdiagnoses. This paper introduces PatchCTG, a transformer-based model specifically designed for CTG analysis, employing patch-based tokenisation, instance normalisation and channel-independent processing to capture essential local and global temporal dependencies within CTG signals. PatchCTG was evaluated on the Oxford Maternity (OXMAT) dataset, comprising over 20,000 CTG traces across diverse clinical outcomes after applying the inclusion and exclusion criteria. With extensive hyperparameter optimisation, PatchCTG achieved an AUC of 77%, with specificity of 88% and sensitivity of 57% at Youden's index threshold, demonstrating adaptability to various clinical needs. Testing across varying temporal thresholds showed robust predictive performance, particularly with finetuning on data closer to delivery, achieving a sensitivity of 52% and specificity of 88% for near-delivery cases. These findings suggest the potential of PatchCTG to enhance clinical decision-making in antepartum care by providing a reliable, objective tool for fetal health assessment. The source code is available at https://github.com/jaleedkhan/PatchCTG.
The OxMat dataset: a multimodal resource for the development of AI-driven technologies in maternal and newborn child health
Apr 11, 2024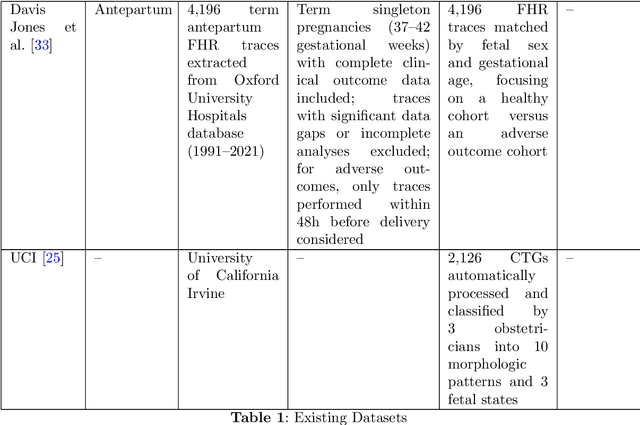
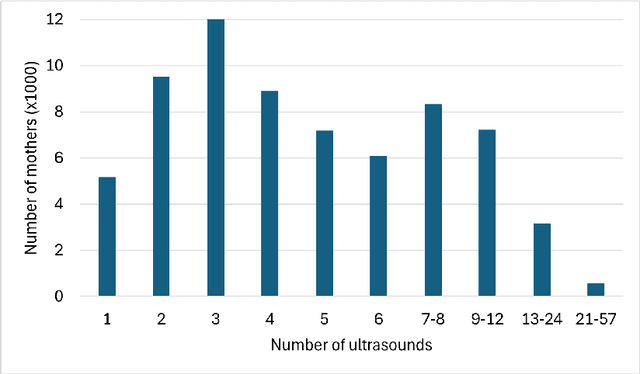
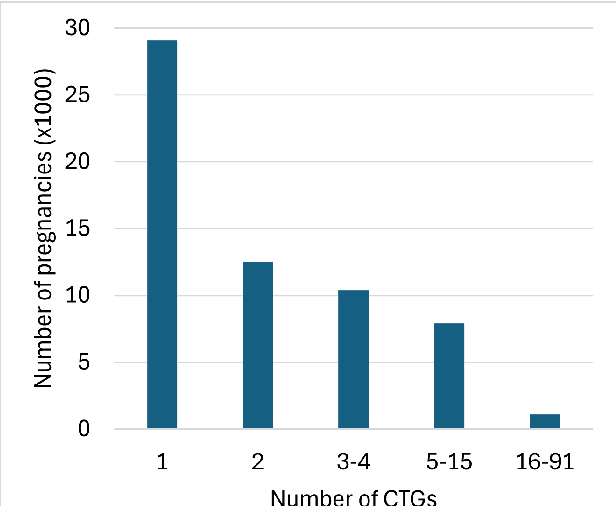
Abstract:The rapid advancement of Artificial Intelligence (AI) in healthcare presents a unique opportunity for advancements in obstetric care, particularly through the analysis of cardiotocography (CTG) for fetal monitoring. However, the effectiveness of such technologies depends upon the availability of large, high-quality datasets that are suitable for machine learning. This paper introduces the Oxford Maternity (OxMat) dataset, the world's largest curated dataset of CTGs, featuring raw time series CTG data and extensive clinical data for both mothers and babies, which is ideally placed for machine learning. The OxMat dataset addresses the critical gap in women's health data by providing over 177,211 unique CTG recordings from 51,036 pregnancies, carefully curated and reviewed since 1991. The dataset also comprises over 200 antepartum, intrapartum and postpartum clinical variables, ensuring near-complete data for crucial outcomes such as stillbirth and acidaemia. While this dataset also covers the intrapartum stage, around 94% of the constituent CTGS are antepartum. This allows for a unique focus on the underserved antepartum period, in which early detection of at-risk fetuses can significantly improve health outcomes. Our comprehensive review of existing datasets reveals the limitations of current datasets: primarily, their lack of sufficient volume, detailed clinical data and antepartum data. The OxMat dataset lays a foundation for future AI-driven prenatal care, offering a robust resource for developing and testing algorithms aimed at improving maternal and fetal health outcomes.
Towards Deep Cellular Phenotyping in Placental Histology
May 25, 2018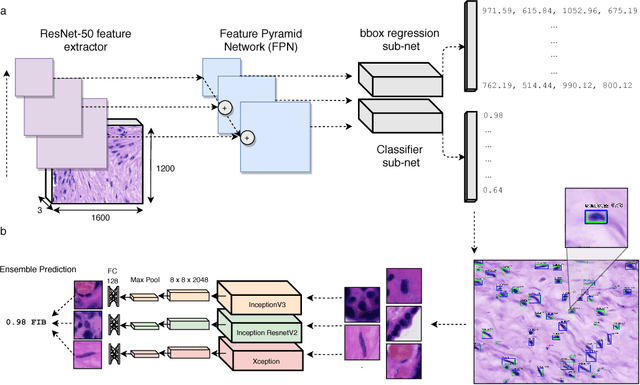
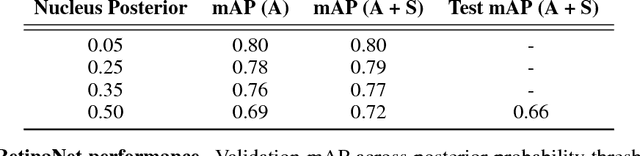
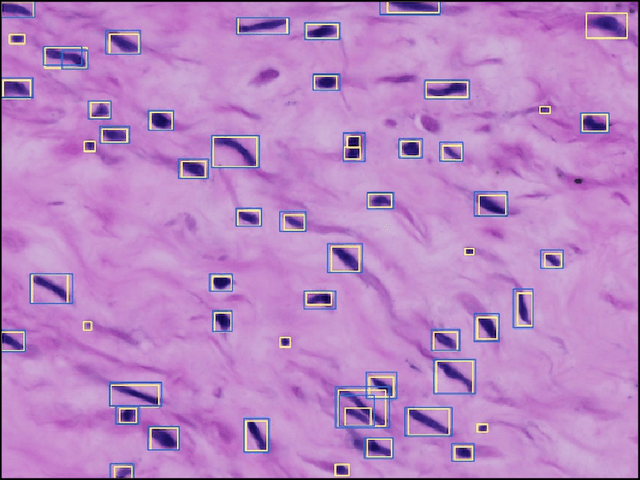
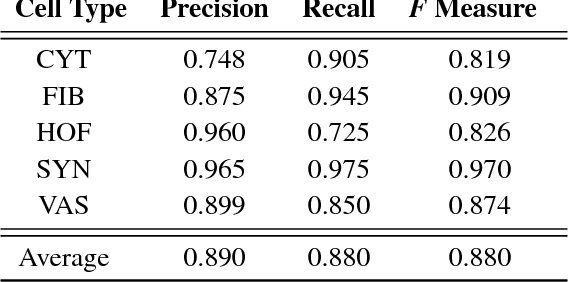
Abstract:The placenta is a complex organ, playing multiple roles during fetal development. Very little is known about the association between placental morphological abnormalities and fetal physiology. In this work, we present an open sourced, computationally tractable deep learning pipeline to analyse placenta histology at the level of the cell. By utilising two deep Convolutional Neural Network architectures and transfer learning, we can robustly localise and classify placental cells within five classes with an accuracy of 89%. Furthermore, we learn deep embeddings encoding phenotypic knowledge that is capable of both stratifying five distinct cell populations and learn intraclass phenotypic variance. We envisage that the automation of this pipeline to population scale studies of placenta histology has the potential to improve our understanding of basic cellular placental biology and its variations, particularly its role in predicting adverse birth outcomes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge