Manoj Agarwal
Biomarker Gene Identification for Breast Cancer Classification
Nov 29, 2021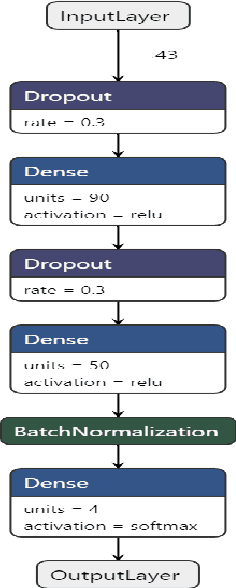
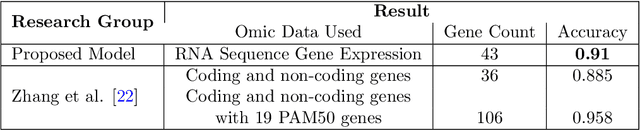
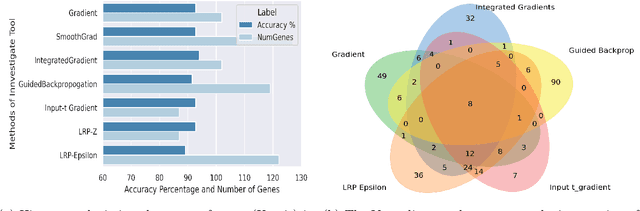
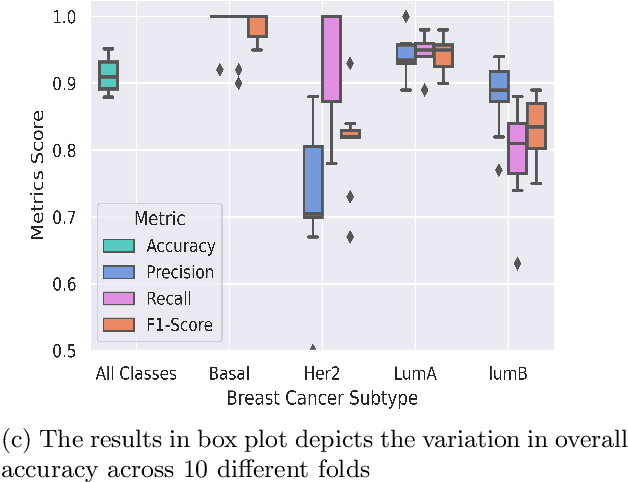
Abstract:BACKGROUND: Breast cancer has emerged as one of the most prevalent cancers among women leading to a high mortality rate. Due to the heterogeneous nature of breast cancer, there is a need to identify differentially expressed genes associated with breast cancer subtypes for its timely diagnosis and treatment. OBJECTIVE: To identify a small gene set for each of the four breast cancer subtypes that could act as its signature, the paper proposes a novel algorithm for gene signature identification. METHODS: The present work uses interpretable AI methods to investigate the predictions made by the deep neural network employed for subtype classification to identify biomarkers using the TCGA breast cancer RNA Sequence data. RESULTS: The proposed algorithm led to the discovery of a set of 43 differentially expressed gene signatures. We achieved a competitive average 10-fold accuracy of 0.91, using neural network classifier. Further, gene set analysis revealed several relevant pathways, such as GRB7 events in ERBB2 and p53 signaling pathway. Using the Pearson correlation matrix, we noted that the subtype-specific genes are correlated within each subtype. CONCLUSIONS: The proposed technique enables us to find a concise and clinically relevant gene signature set.
Deep Learning Based Model for Breast Cancer Subtype Classification
Nov 09, 2021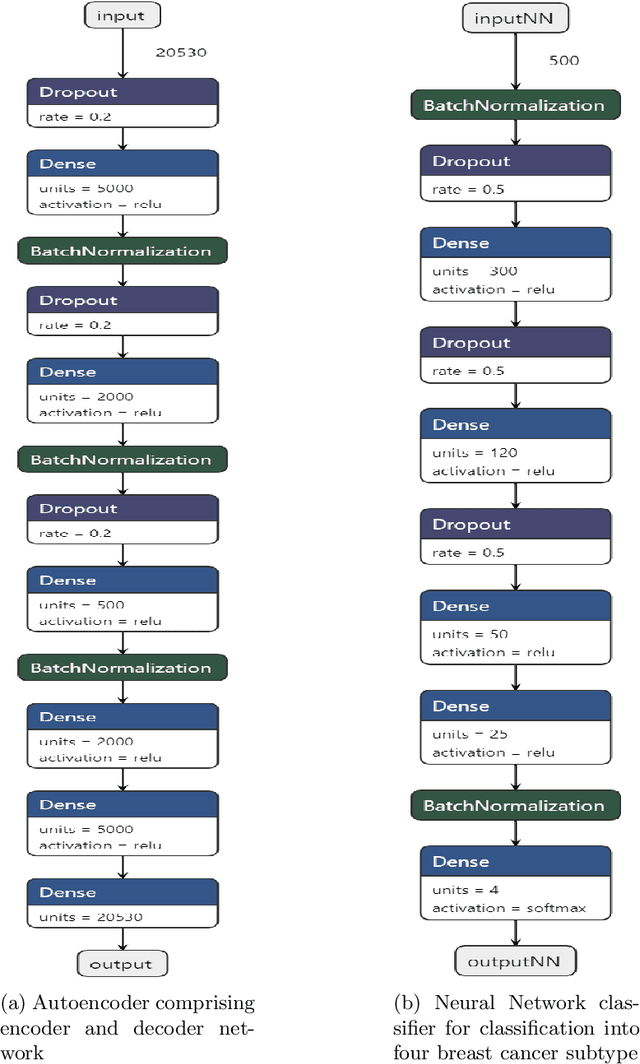
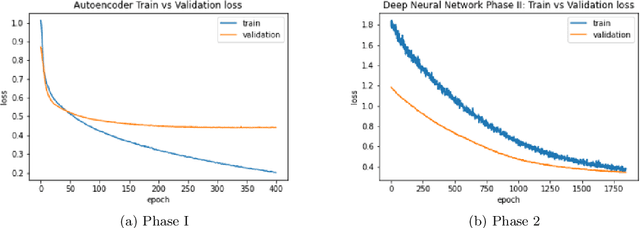
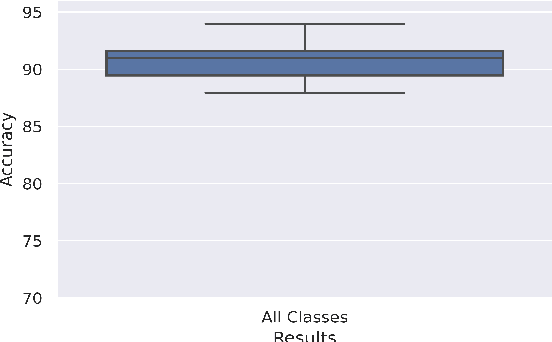
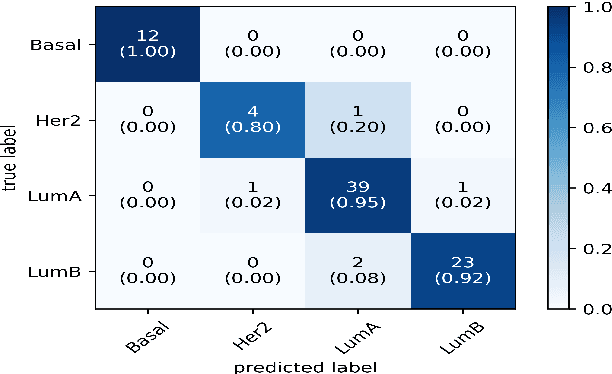
Abstract:Breast cancer has long been a prominent cause of mortality among women. Diagnosis, therapy, and prognosis are now possible, thanks to the availability of RNA sequencing tools capable of recording gene expression data. Molecular subtyping being closely related to devising clinical strategy and prognosis, this paper focuses on the use of gene expression data for the classification of breast cancer into four subtypes, namely, Basal, Her2, LumA, and LumB. In stage 1, we suggested a deep learning-based model that uses an autoencoder to reduce dimensionality. The size of the feature set is reduced from 20,530 gene expression values to 500 by using an autoencoder. This encoded representation is passed to the deep neural network of the second stage for the classification of patients into four molecular subtypes of breast cancer. By deploying the combined network of stages 1 and 2, we have been able to attain a mean 10-fold test accuracy of 0.907 on the TCGA breast cancer dataset. The proposed framework is fairly robust throughout 10 different runs, as shown by the boxplot for classification accuracy. Compared to related work reported in the literature, we have achieved a competitive outcome. In conclusion, the proposed two-stage deep learning-based model is able to accurately classify four breast cancer subtypes, highlighting the autoencoder's capacity to deduce the compact representation and the neural network classifier's ability to correctly label breast cancer patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge