Lynn E. Eberly
Multiway sparse distance weighted discrimination
Oct 11, 2021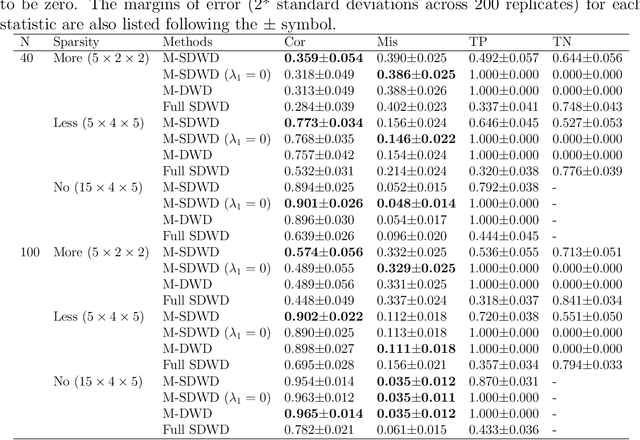
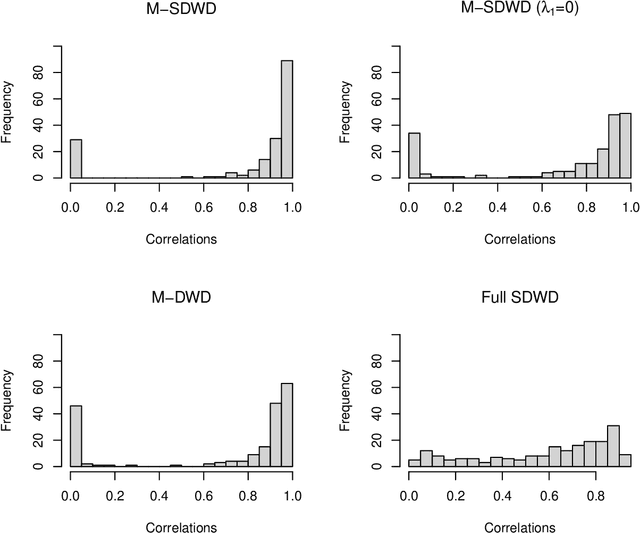
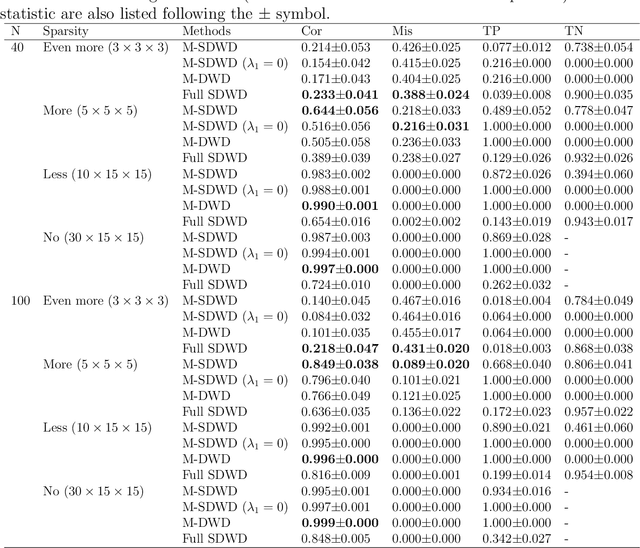
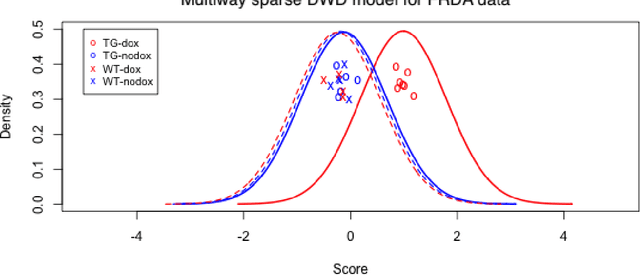
Abstract:Modern data often take the form of a multiway array. However, most classification methods are designed for vectors, i.e., 1-way arrays. Distance weighted discrimination (DWD) is a popular high-dimensional classification method that has been extended to the multiway context, with dramatic improvements in performance when data have multiway structure. However, the previous implementation of multiway DWD was restricted to classification of matrices, and did not account for sparsity. In this paper, we develop a general framework for multiway classification which is applicable to any number of dimensions and any degree of sparsity. We conducted extensive simulation studies, showing that our model is robust to the degree of sparsity and improves classification accuracy when the data have multiway structure. For our motivating application, magnetic resonance spectroscopy (MRS) was used to measure the abundance of several metabolites across multiple neurological regions and across multiple time points in a mouse model of Friedreich's ataxia, yielding a four-way data array. Our method reveals a robust and interpretable multi-region metabolomic signal that discriminates the groups of interest. We also successfully apply our method to gene expression time course data for multiple sclerosis treatment. An R implementation is available in the package MultiwayClassification at http://github.com/lockEF/MultiwayClassification .
Discriminating sample groups with multi-way data
Jun 26, 2016



Abstract:High-dimensional linear classifiers, such as the support vector machine (SVM) and distance weighted discrimination (DWD), are commonly used in biomedical research to distinguish groups of subjects based on a large number of features. However, their use is limited to applications where a single vector of features is measured for each subject. In practice data are often multi-way, or measured over multiple dimensions. For example, metabolite abundance may be measured over multiple regions or tissues, or gene expression may be measured over multiple time points, for the same subjects. We propose a framework for linear classification of high-dimensional multi-way data, in which coefficients can be factorized into weights that are specific to each dimension. More generally, the coefficients for each measurement in a multi-way dataset are assumed to have low-rank structure. This framework extends existing classification techniques, and we have implemented multi-way versions of SVM and DWD. We describe informative simulation results, and apply multi-way DWD to data for two very different clinical research studies. The first study uses metabolite magnetic resonance spectroscopy data over multiple brain regions to compare patients with and without spinocerebellar ataxia, the second uses publicly available gene expression time-course data to compare treatment responses for patients with multiple sclerosis. Our method improves performance and simplifies interpretation over naive applications of full rank linear classification to multi-way data. An R package is available at https://github.com/lockEF/MultiwayClassification .
* 25 pages, 5 figures, 2 tables
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge