Lydia Neary- Zajiczek
Deep Learning Enhanced Extended Depth-of-Field for Thick Blood-Film Malaria High-Throughput Microscopy
Jun 18, 2019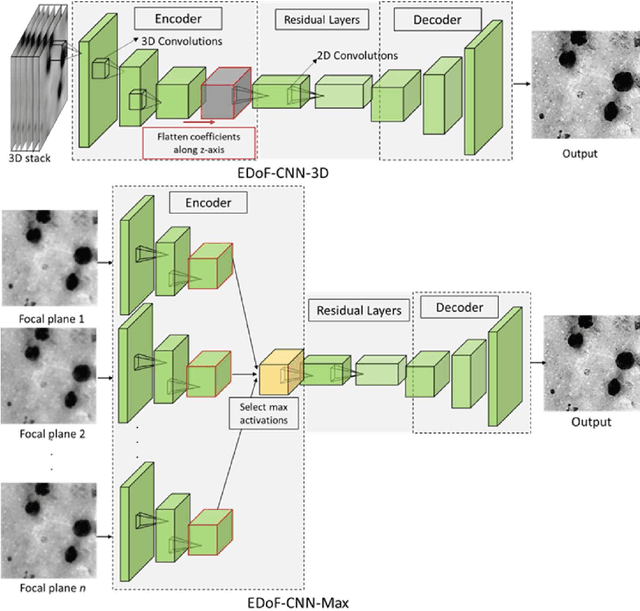
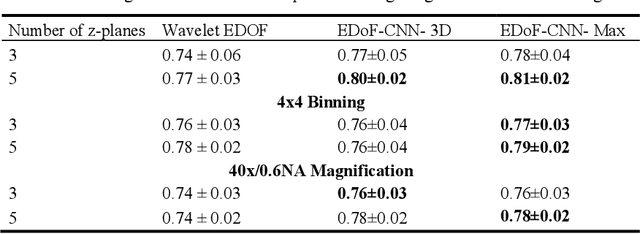
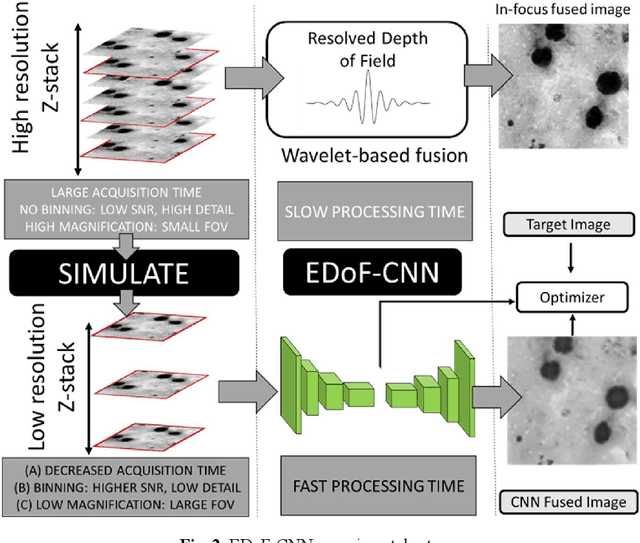
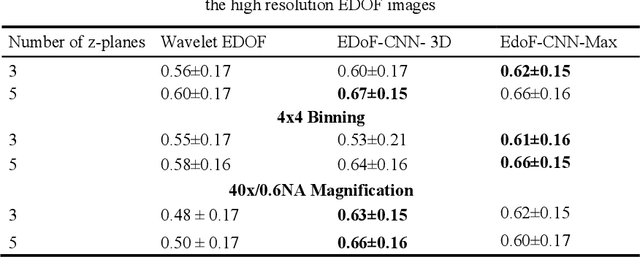
Abstract:Fast accurate diagnosis of malaria is still a global health challenge for which automated digital-pathology approaches could provide scalable solutions amenable to be deployed in low-to-middle income countries. Here we address the problem of Extended Depth-of-Field (EDoF) in thick blood film microscopy for rapid automated malaria diagnosis. High magnification oil-objectives (100x) with large numerical aperture are usually preferred to resolve the fine structural details that help separate true parasites from distractors. However, such objectives have a very limited depth-of-field requiring the acquisition of a series of images at different focal planes per field of view (FOV). Current EDoF techniques based on multi-scale decompositions are time consuming and therefore not suited for high-throughput analysis of specimens. To overcome this challenge, we developed a new deep learning method based on Convolutional Neural Networks (EDoF-CNN) that is able to rapidly perform the extended depth-of-field while also enhancing the spatial resolution of the resulting fused image. We evaluated our approach using simulated low-resolution z-stacks from Giemsa-stained thick blood smears from patients presenting with Plasmodium falciparum malaria. The EDoF-CNN allows speed-up of our digital-pathology acquisition platform and significantly improves the quality of the EDoF compared to the traditional multi-scaled approaches when applied to lower resolution stacks corresponding to acquisitions with fewer focal planes, large camera pixel binning or lower magnification objectives (larger FOV). We use the parasite detection accuracy of a deep learning model on the EDoFs as a concrete, task-specific measure of performance of this approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge