Lucas Ferrari de Oliveira
Optimizing Neural Architecture Search using Limited GPU Time in a Dynamic Search Space: A Gene Expression Programming Approach
May 15, 2020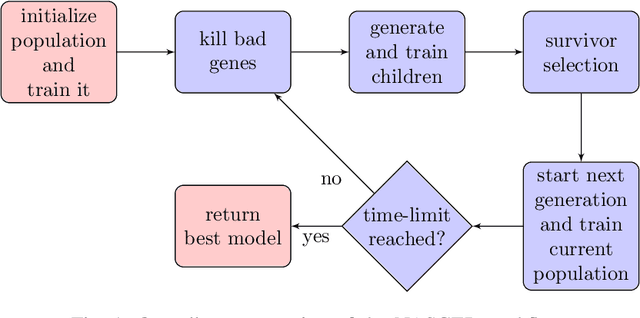
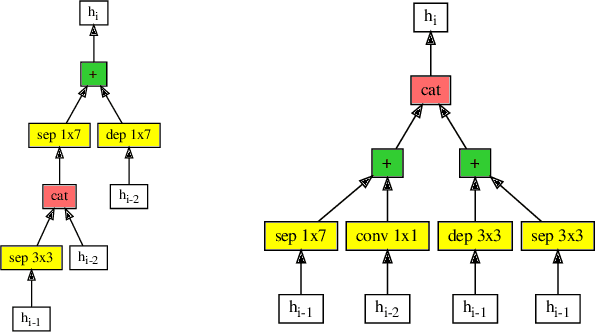
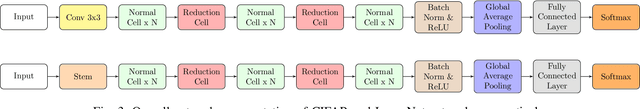
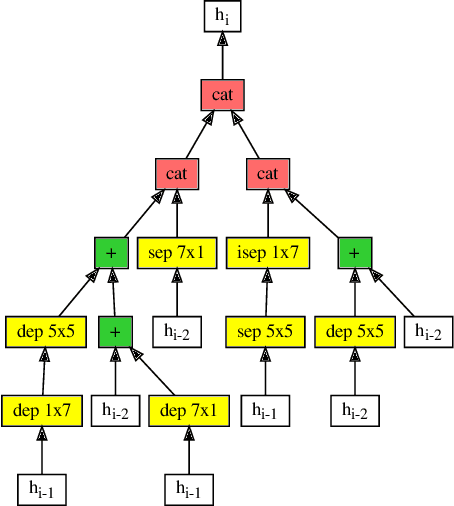
Abstract:Efficient identification of people and objects, segmentation of regions of interest and extraction of relevant data in images, texts, audios and videos are evolving considerably in these past years, which deep learning methods, combined with recent improvements in computational resources, contributed greatly for this achievement. Although its outstanding potential, development of efficient architectures and modules requires expert knowledge and amount of resource time available. In this paper, we propose an evolutionary-based neural architecture search approach for efficient discovery of convolutional models in a dynamic search space, within only 24 GPU hours. With its efficient search environment and phenotype representation, Gene Expression Programming is adapted for network's cell generation. Despite having limited GPU resource time and broad search space, our proposal achieved similar state-of-the-art to manually-designed convolutional networks and also NAS-generated ones, even beating similar constrained evolutionary-based NAS works. The best cells in different runs achieved stable results, with a mean error of 2.82% in CIFAR-10 dataset (which the best model achieved an error of 2.67%) and 18.83% for CIFAR-100 (best model with 18.16%). For ImageNet in the mobile setting, our best model achieved top-1 and top-5 errors of 29.51% and 10.37%, respectively. Although evolutionary-based NAS works were reported to require a considerable amount of GPU time for architecture search, our approach obtained promising results in little time, encouraging further experiments in evolutionary-based NAS, for search and network representation improvements.
Extracting Lungs from CT Images using Fully Convolutional Networks
Apr 27, 2018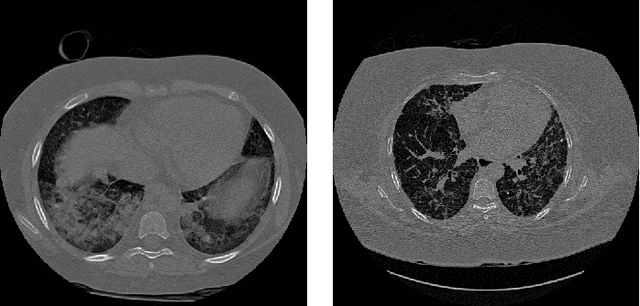
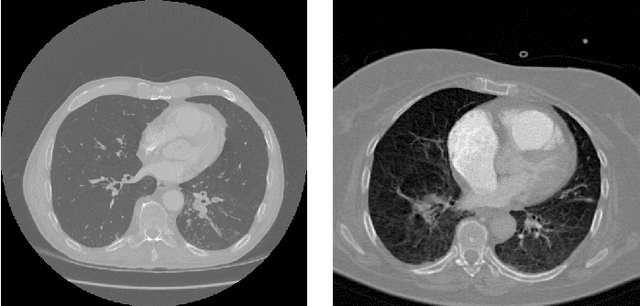
Abstract:Analysis of cancer and other pathological diseases, like the interstitial lung diseases (ILDs), is usually possible through Computed Tomography (CT) scans. To aid this, a preprocessing step of segmentation is performed to reduce the area to be analyzed, segmenting the lungs and removing unimportant regions. Generally, complex methods are developed to extract the lung region, also using hand-made feature extractors to enhance segmentation. With the popularity of deep learning techniques and its automated feature learning, we propose a lung segmentation approach using fully convolutional networks (FCNs) combined with fully connected conditional random fields (CRF), employed in many state-of-the-art segmentation works. Aiming to develop a generalized approach, the publicly available datasets from University Hospitals of Geneva (HUG) and VESSEL12 challenge were studied, including many healthy and pathological CT scans for evaluation. Experiments using the dataset individually, its trained model on the other dataset and a combination of both datasets were employed. Dice scores of $98.67\%\pm0.94\%$ for the HUG-ILD dataset and $99.19\%\pm0.37\%$ for the VESSEL12 dataset were achieved, outperforming works in the former and obtaining similar state-of-the-art results in the latter dataset, showing the capability in using deep learning approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge