Lorenzo Bini
LapDDPM: A Conditional Graph Diffusion Model for scRNA-seq Generation with Spectral Adversarial Perturbations
Jun 16, 2025Abstract:Generating high-fidelity and biologically plausible synthetic single-cell RNA sequencing (scRNA-seq) data, especially with conditional control, is challenging due to its high dimensionality, sparsity, and complex biological variations. Existing generative models often struggle to capture these unique characteristics and ensure robustness to structural noise in cellular networks. We introduce LapDDPM, a novel conditional Graph Diffusion Probabilistic Model for robust and high-fidelity scRNA-seq generation. LapDDPM uniquely integrates graph-based representations with a score-based diffusion model, enhanced by a novel spectral adversarial perturbation mechanism on graph edge weights. Our contributions are threefold: we leverage Laplacian Positional Encodings (LPEs) to enrich the latent space with crucial cellular relationship information; we develop a conditional score-based diffusion model for effective learning and generation from complex scRNA-seq distributions; and we employ a unique spectral adversarial training scheme on graph edge weights, boosting robustness against structural variations. Extensive experiments on diverse scRNA-seq datasets demonstrate LapDDPM's superior performance, achieving high fidelity and generating biologically-plausible, cell-type-specific samples. LapDDPM sets a new benchmark for conditional scRNA-seq data generation, offering a robust tool for various downstream biological applications.
Characterizing Massive Activations of Attention Mechanism in Graph Neural Networks
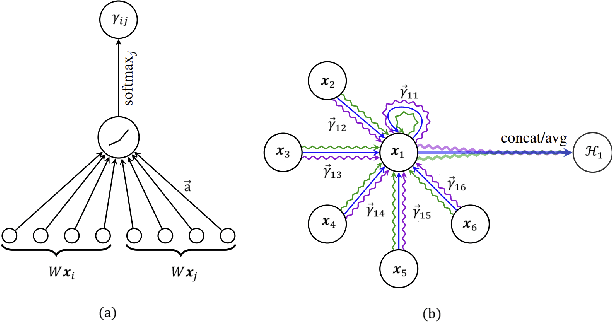
Sep 05, 2024Abstract:Graph Neural Networks (GNNs) have become increasingly popular for effectively modeling data with graph structures. Recently, attention mechanisms have been integrated into GNNs to improve their ability to capture complex patterns. This paper presents the first comprehensive study revealing a critical, unexplored consequence of this integration: the emergence of Massive Activations (MAs) within attention layers. We introduce a novel method for detecting and analyzing MAs, focusing on edge features in different graph transformer architectures. Our study assesses various GNN models using benchmark datasets, including ZINC, TOX21, and PROTEINS. Key contributions include (1) establishing the direct link between attention mechanisms and MAs generation in GNNs, (2) developing a robust definition and detection method for MAs based on activation ratio distributions, (3) introducing the Explicit Bias Term (EBT) as a potential countermeasure and exploring it as an adversarial framework to assess models robustness based on the presence or absence of MAs. Our findings highlight the prevalence and impact of attention-induced MAs across different architectures, such as GraphTransformer, GraphiT, and SAN. The study reveals the complex interplay between attention mechanisms, model architecture, dataset characteristics, and MAs emergence, providing crucial insights for developing more robust and reliable graph models.
Injecting Hierarchical Biological Priors into Graph Neural Networks for Flow Cytometry Prediction
May 28, 2024Abstract:In the complex landscape of hematologic samples such as peripheral blood or bone marrow derived from flow cytometry (FC) data, cell-level prediction presents profound challenges. This work explores injecting hierarchical prior knowledge into graph neural networks (GNNs) for single-cell multi-class classification of tabular cellular data. By representing the data as graphs and encoding hierarchical relationships between classes, we propose our hierarchical plug-in method to be applied to several GNN models, namely, FCHC-GNN, and effectively designed to capture neighborhood information crucial for single-cell FC domain. Extensive experiments on our cohort of 19 distinct patients, demonstrate that incorporating hierarchical biological constraints boosts performance significantly across multiple metrics compared to baseline GNNs without such priors. The proposed approach highlights the importance of structured inductive biases for gaining improved generalization in complex biological prediction tasks.
HemaGraph: Breaking Barriers in Hematologic Single Cell Classification with Graph Attention
Feb 28, 2024Abstract:In the realm of hematologic cell populations classification, the intricate patterns within flow cytometry data necessitate advanced analytical tools. This paper presents 'HemaGraph', a novel framework based on Graph Attention Networks (GATs) for single-cell multi-class classification of hematological cells from flow cytometry data. Harnessing the power of GATs, our method captures subtle cell relationships, offering highly accurate patient profiling. Based on evaluation of data from 30 patients, HemaGraph demonstrates classification performance across five different cell classes, outperforming traditional methodologies and state-of-the-art methods. Moreover, the uniqueness of this framework lies in the training and testing phase of HemaGraph, where it has been applied for extremely large graphs, containing up to hundreds of thousands of nodes and two million edges, to detect low frequency cell populations (e.g. 0.01% for one population), with accuracies reaching 98%. Our findings underscore the potential of HemaGraph in improving hematoligic multi-class classification, paving the way for patient-personalized interventions. To the best of our knowledge, this is the first effort to use GATs, and Graph Neural Networks (GNNs) in general, to classify cell populations from single-cell flow cytometry data. We envision applying this method to single-cell data from larger cohort of patients and on other hematologic diseases.
Why Attention Graphs Are All We Need: Pioneering Hierarchical Classification of Hematologic Cell Populations with LeukoGraph
Feb 28, 2024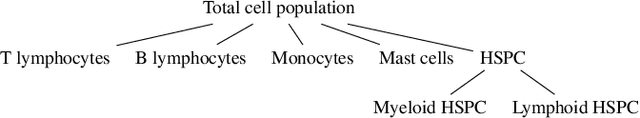
Abstract:In the complex landscape of hematologic samples such as peripheral blood or bone marrow, cell classification, delineating diverse populations into a hierarchical structure, presents profound challenges. This study presents LeukoGraph, a recently developed framework designed explicitly for this purpose employing graph attention networks (GATs) to navigate hierarchical classification (HC) complexities. Notably, LeukoGraph stands as a pioneering effort, marking the application of graph neural networks (GNNs) for hierarchical inference on graphs, accommodating up to one million nodes and millions of edges, all derived from flow cytometry data. LeukoGraph intricately addresses a classification paradigm where for example four different cell populations undergo flat categorization, while a fifth diverges into two distinct child branches, exemplifying the nuanced hierarchical structure inherent in complex datasets. The technique is more general than this example. A hallmark achievement of LeukoGraph is its F-score of 98%, significantly outclassing prevailing state-of-the-art methodologies. Crucially, LeukoGraph's prowess extends beyond theoretical innovation, showcasing remarkable precision in predicting both flat and hierarchical cell types across flow cytometry datasets from 30 distinct patients. This precision is further underscored by LeukoGraph's ability to maintain a correct label ratio, despite the inherent challenges posed by hierarchical classifications.
FlowCyt: A Comparative Study of Deep Learning Approaches for Multi-Class Classification in Flow Cytometry Benchmarking
Feb 28, 2024



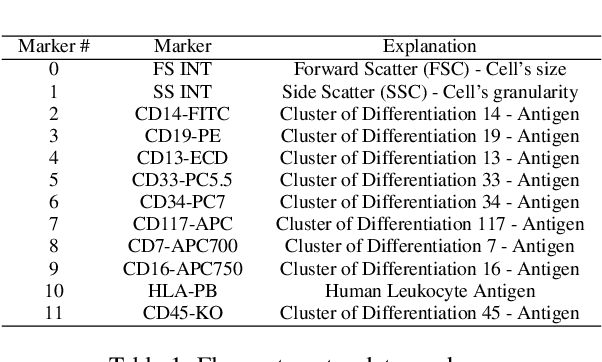
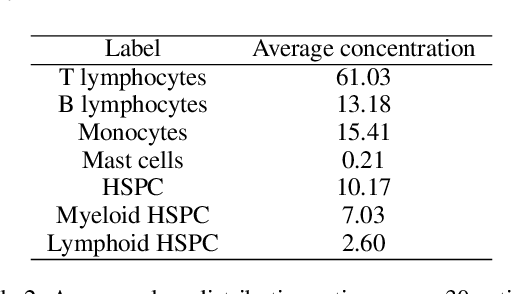
Abstract:This paper presents FlowCyt, the first comprehensive benchmark for multi-class single-cell classification in flow cytometry data. The dataset comprises bone marrow samples from 30 patients, with each cell characterized by twelve markers. Ground truth labels identify five hematological cell types: T lymphocytes, B lymphocytes, Monocytes, Mast cells, and Hematopoietic Stem/Progenitor Cells (HSPCs). Experiments utilize supervised inductive learning and semi-supervised transductive learning on up to 1 million cells per patient. Baseline methods include Gaussian Mixture Models, XGBoost, Random Forests, Deep Neural Networks, and Graph Neural Networks (GNNs). GNNs demonstrate superior performance by exploiting spatial relationships in graph-encoded data. The benchmark allows standardized evaluation of clinically relevant classification tasks, along with exploratory analyses to gain insights into hematological cell phenotypes. This represents the first public flow cytometry benchmark with a richly annotated, heterogeneous dataset. It will empower the development and rigorous assessment of novel methodologies for single-cell analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge