Lipeng Lai
siDPT: siRNA Efficacy Prediction via Debiased Preference-Pair Transformer
Sep 19, 2025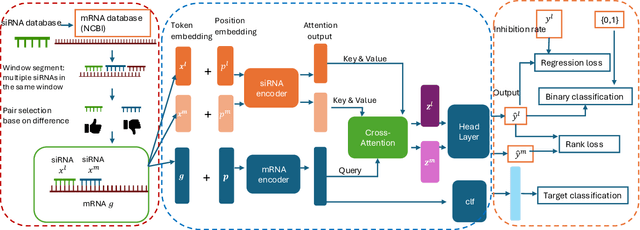
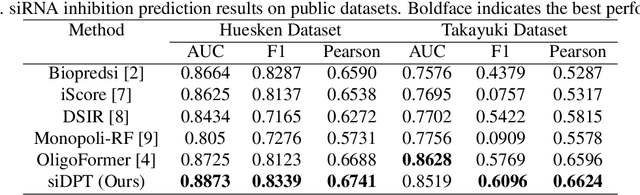
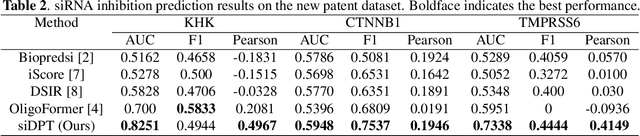
Abstract:Small interfering RNA (siRNA) is a short double-stranded RNA molecule (about 21-23 nucleotides) with the potential to cure diseases by silencing the function of target genes. Due to its well-understood mechanism, many siRNA-based drugs have been evaluated in clinical trials. However, selecting effective binding regions and designing siRNA sequences requires extensive experimentation, making the process costly. As genomic resources and publicly available siRNA datasets continue to grow, data-driven models can be leveraged to better understand siRNA-mRNA interactions. To fully exploit such data, curating high-quality siRNA datasets is essential to minimize experimental errors and noise. We propose siDPT: siRNA efficacy Prediction via Debiased Preference-Pair Transformer, a framework that constructs a preference-pair dataset and designs an siRNA-mRNA interactive transformer with debiased ranking objectives to improve siRNA inhibition prediction and generalization. We evaluate our approach using two public datasets and one newly collected patent dataset. Our model demonstrates substantial improvement in Pearson correlation and strong performance across other metrics.
mRNA2vec: mRNA Embedding with Language Model in the 5'UTR-CDS for mRNA Design
Aug 16, 2024Abstract:Messenger RNA (mRNA)-based vaccines are accelerating the discovery of new drugs and revolutionizing the pharmaceutical industry. However, selecting particular mRNA sequences for vaccines and therapeutics from extensive mRNA libraries is costly. Effective mRNA therapeutics require carefully designed sequences with optimized expression levels and stability. This paper proposes a novel contextual language model (LM)-based embedding method: mRNA2vec. In contrast to existing mRNA embedding approaches, our method is based on the self-supervised teacher-student learning framework of data2vec. We jointly use the 5' untranslated region (UTR) and coding sequence (CDS) region as the input sequences. We adapt our LM-based approach specifically to mRNA by 1) considering the importance of location on the mRNA sequence with probabilistic masking, 2) using Minimum Free Energy (MFE) prediction and Secondary Structure (SS) classification as additional pretext tasks. mRNA2vec demonstrates significant improvements in translation efficiency (TE) and expression level (EL) prediction tasks in UTR compared to SOTA methods such as UTR-LM. It also gives a competitive performance in mRNA stability and protein production level tasks in CDS such as CodonBERT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge