Laurent Milot
Comparing Deep Learning strategies for paired but unregistered multimodal segmentation of the liver in T1 and T2-weighted MRI
Jan 18, 2021
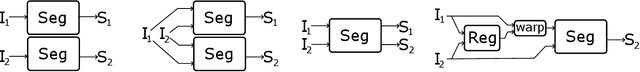
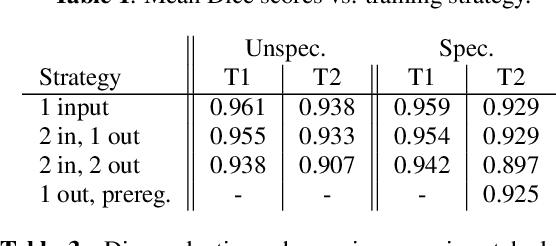
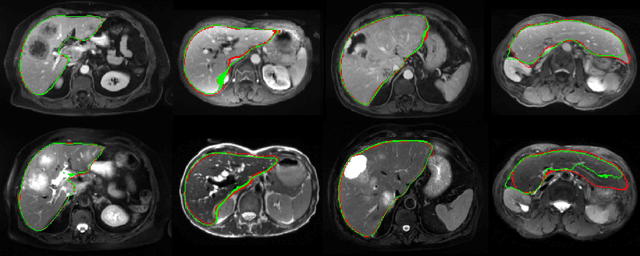
Abstract:We address the problem of multimodal liver segmentation in paired but unregistered T1 and T2-weighted MR images. We compare several strategies described in the literature, with or without multi-task training, with or without pre-registration. We also compare different loss functions (cross-entropy, Dice loss, and three adversarial losses). All methods achieved comparable performances with the exception of a multi-task setting that performs both segmentations at once, which performed poorly.
AMINN: Autoencoder-based Multiple Instance Neural Network for Outcome Prediction of Multifocal Liver Metastases
Dec 12, 2020



Abstract:Colorectal cancer is one of the most common and lethal cancers and colorectal cancer liver metastases (CRLM) is the major cause of death in patients with colorectal cancer. Multifocality occurs frequently in CRLM, but is relatively unexplored in CRLM outcome prediction. Most existing clinical and imaging biomarkers do not take the imaging features of all multifocal lesions into account. In this paper, we present an end-to-end autoencoder-based multiple instance neural network (AMINN) for the prediction of survival outcomes in multifocal CRLM patients using radiomic features extracted from contrast-enhanced MRIs. Specifically, we jointly train an autoencoder to reconstruct input features and a multiple instance network to make predictions by aggregating information from all tumour lesions of a patient. In addition, we incorporate a two-step normalization technique to improve the training of deep neural networks, built on the observation that the distributions of radiomic features are almost always severely skewed. Experimental results empirically validated our hypothesis that incorporating imaging features of all lesions improves outcome prediction for multifocal cancer. The proposed ADMINN framework achieved an area under the ROC curve (AUC) of 0.70, which is 19.5% higher than baseline methods. We built a risk score based on the outputs of our network and compared it to other clinical and imaging biomarkers. Our risk score is the only one that achieved statistical significance in univariate and multivariate cox proportional hazard modeling in our cohort of multifocal CRLM patients. The effectiveness of incorporating all lesions and applying two-step normalization is demonstrated by a series of ablation studies. Our code will be released after the peer-review process.
Unsupervised Clustering of Quantitative Imaging Phenotypes using Autoencoder and Gaussian Mixture Model
Sep 06, 2019



Abstract:Quantitative medical image computing (radiomics) has been widely applied to build prediction models from medical images. However, overfitting is a significant issue in conventional radiomics, where a large number of radiomic features are directly used to train and test models that predict genotypes or clinical outcomes. In order to tackle this problem, we propose an unsupervised learning pipeline composed of an autoencoder for representation learning of radiomic features and a Gaussian mixture model based on minimum message length criterion for clustering. By incorporating probabilistic modeling, disease heterogeneity has been taken into account. The performance of the proposed pipeline was evaluated on an institutional MRI cohort of 108 patients with colorectal cancer liver metastases. Our approach is capable of automatically selecting the optimal number of clusters and assigns patients into clusters (imaging subtypes) with significantly different survival rates. Our method outperforms other unsupervised clustering methods that have been used for radiomics analysis and has comparable performance to a state-of-the-art imaging biomarker.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge