Kyler Larsen
Using Deep Learning to Improve Early Diagnosis of Pneumonia in Underdeveloped Countries
Oct 10, 2022
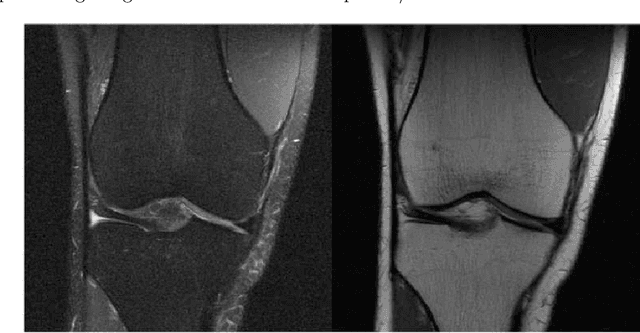
Abstract:As advancements in technology and medicine are being made, many countries are still unable to access quality medical care due to cost and lack of qualified medical personnel. This discrepancy in healthcare has caused many preventable deaths, either due to lack of detection or lack of care. One of the most prevalent diseases in the world is pneumonia, an infection of the lungs that killed 2.56 million people worldwide in 2017. In this same year, the United States recorded a pneumonia death rate of 15.88 people per 100000 in population, while much of Sub-Saharan Africa, such as Chad and Guinea, experienced death rates of over 150 people per 100000. In sub-Saharan Africa, there is an extreme shortage of doctors and nurses, estimated to be around 2.4 million. The hypothesis being tested is that a deep learning model can receive input in the form of an x-ray and produce a diagnosis with the equivalent accuracy of a physician, compared to a prediagnosed image. The model used in this project is a modified convolutional neural network. The model was trained on a set of 2000 x-ray images that have predetermined normal and abnormal lung findings, and then tested on a set of 400 images that contains evenly split images of pneumonia and healthy lungs. For each computer-run test, data was collected on a base measurement of accuracy, as well as more specific metrics such as specificity and sensitivity. Results show that the algorithm tested was able to accurately identify abnormal lung findings an average of 82.5% of the time. The model achieved a maximum specificity of 98.5% and a maximum sensitivity of 90% separately, and the highest simultaneous values of these two metrics was a sensitivity of 90% and a specificity of 78.5%. This research can be further improved by testing other deep learning models as well as machine learning models to improve the metric scores and chance of correct diagnoses.
A Deep Learning Approach Using Masked Image Modeling for Reconstruction of Undersampled K-spaces
Aug 24, 2022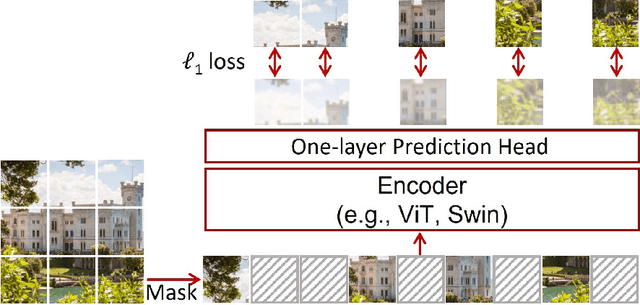
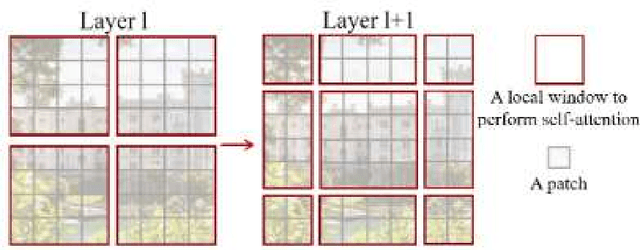
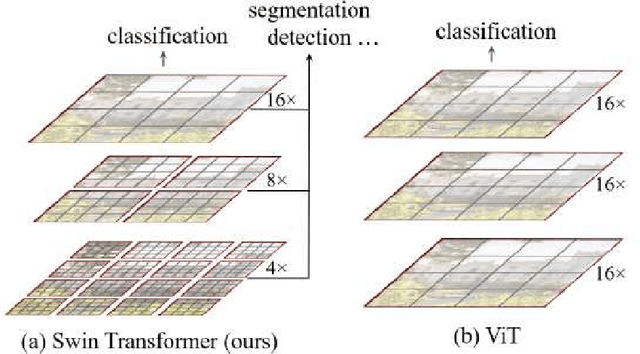
Abstract:Magnetic Resonance Imaging (MRI) scans are time consuming and precarious, since the patients remain still in a confined space for extended periods of time. To reduce scanning time, some experts have experimented with undersampled k spaces, trying to use deep learning to predict the fully sampled result. These studies report that as many as 20 to 30 minutes could be saved off a scan that takes an hour or more. However, none of these studies have explored the possibility of using masked image modeling (MIM) to predict the missing parts of MRI k spaces. This study makes use of 11161 reconstructed MRI and k spaces of knee MRI images from Facebook's fastmri dataset. This tests a modified version of an existing model using baseline shifted window (Swin) and vision transformer architectures that makes use of MIM on undersampled k spaces to predict the full k space and consequently the full MRI image. Modifications were made using pytorch and numpy libraries, and were published to a github repository. After the model reconstructed the k space images, the basic Fourier transform was applied to determine the actual MRI image. Once the model reached a steady state, experimentation with hyperparameters helped to achieve pinpoint accuracy for the reconstructed images. The model was evaluated through L1 loss, gradient normalization, and structural similarity values. The model produced reconstructed images with L1 loss values averaging to <0.01 and gradient normalization values <0.1 after training finished. The reconstructed k spaces yielded structural similarity values of over 99% for both training and validation with the fully sampled k spaces, while validation loss continually decreased under 0.01. These data strongly support the idea that the algorithm works for MRI reconstruction, as they indicate the model's reconstructed image aligns extremely well with the original, fully sampled k space.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge