Kunal Suri
SuryaKiran at MEDIQA-Sum 2023: Leveraging LoRA for Clinical Dialogue Summarization
Jul 11, 2023
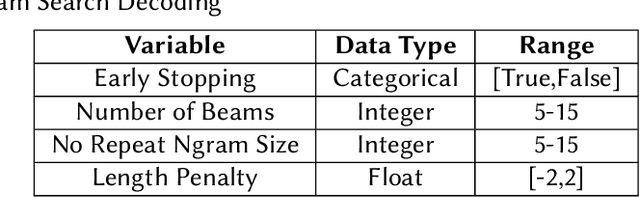
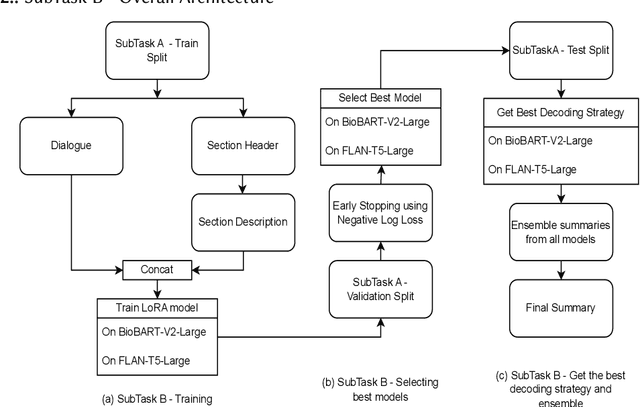
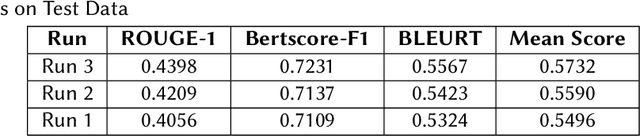
Abstract:Finetuning Large Language Models helps improve the results for domain-specific use cases. End-to-end finetuning of large language models is time and resource intensive and has high storage requirements to store the finetuned version of the large language model. Parameter Efficient Fine Tuning (PEFT) methods address the time and resource challenges by keeping the large language model as a fixed base and add additional layers, which the PEFT methods finetune. This paper demonstrates the evaluation results for one such PEFT method Low Rank Adaptation (LoRA), for Clinical Dialogue Summarization. The evaluation results show that LoRA works at par with end-to-end finetuning for a large language model. The paper presents the evaluations done for solving both the Subtask A and B from ImageCLEFmedical {https://www.imageclef.org/2023/medical}
Language Models sounds the Death Knell of Knowledge Graphs
Jan 10, 2023



Abstract:Healthcare domain generates a lot of unstructured and semi-structured text. Natural Language processing (NLP) has been used extensively to process this data. Deep Learning based NLP especially Large Language Models (LLMs) such as BERT have found broad acceptance and are used extensively for many applications. A Language Model is a probability distribution over a word sequence. Self-supervised Learning on a large corpus of data automatically generates deep learning-based language models. BioBERT and Med-BERT are language models pre-trained for the healthcare domain. Healthcare uses typical NLP tasks such as question answering, information extraction, named entity recognition, and search to simplify and improve processes. However, to ensure robust application of the results, NLP practitioners need to normalize and standardize them. One of the main ways of achieving normalization and standardization is the use of Knowledge Graphs. A Knowledge Graph captures concepts and their relationships for a specific domain, but their creation is time-consuming and requires manual intervention from domain experts, which can prove expensive. SNOMED CT (Systematized Nomenclature of Medicine -- Clinical Terms), Unified Medical Language System (UMLS), and Gene Ontology (GO) are popular ontologies from the healthcare domain. SNOMED CT and UMLS capture concepts such as disease, symptoms and diagnosis and GO is the world's largest source of information on the functions of genes. Healthcare has been dealing with an explosion in information about different types of drugs, diseases, and procedures. This paper argues that using Knowledge Graphs is not the best solution for solving problems in this domain. We present experiments using LLMs for the healthcare domain to demonstrate that language models provide the same functionality as knowledge graphs, thereby making knowledge graphs redundant.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge