Konstantin I. Popov
Utilizing Low-Dimensional Molecular Embeddings for Rapid Chemical Similarity Search
Feb 12, 2024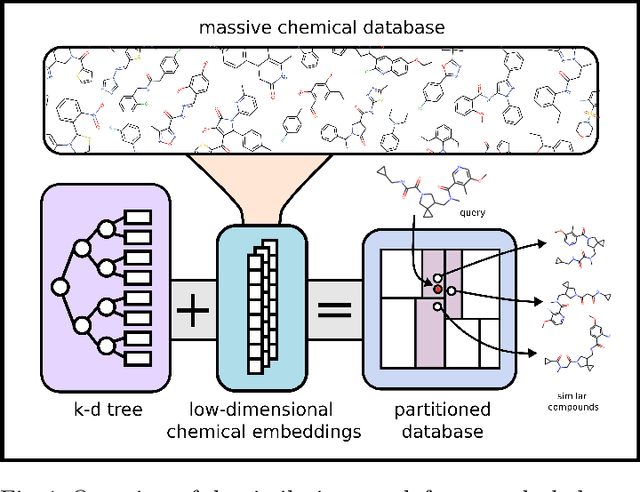
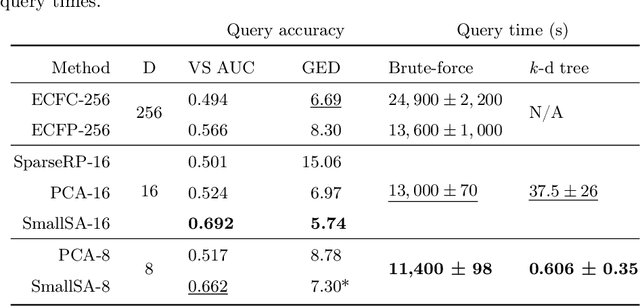
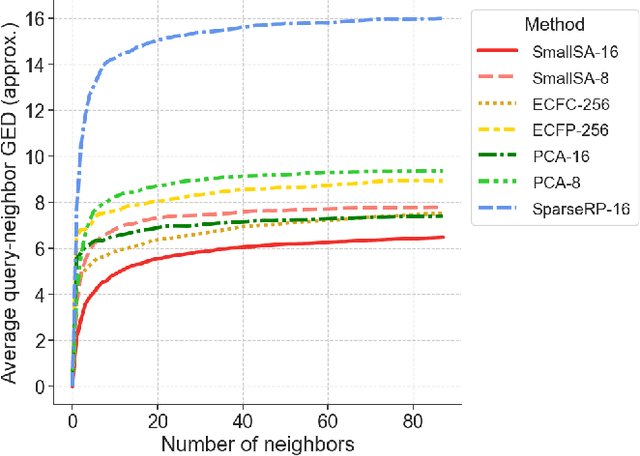
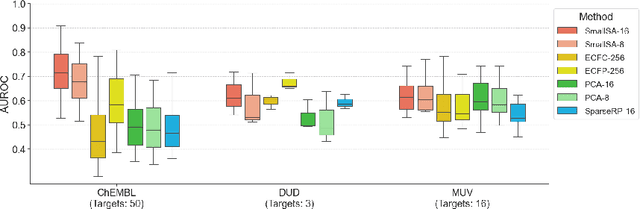
Abstract:Nearest neighbor-based similarity searching is a common task in chemistry, with notable use cases in drug discovery. Yet, some of the most commonly used approaches for this task still leverage a brute-force approach. In practice this can be computationally costly and overly time-consuming, due in part to the sheer size of modern chemical databases. Previous computational advancements for this task have generally relied on improvements to hardware or dataset-specific tricks that lack generalizability. Approaches that leverage lower-complexity searching algorithms remain relatively underexplored. However, many of these algorithms are approximate solutions and/or struggle with typical high-dimensional chemical embeddings. Here we evaluate whether a combination of low-dimensional chemical embeddings and a k-d tree data structure can achieve fast nearest neighbor queries while maintaining performance on standard chemical similarity search benchmarks. We examine different dimensionality reductions of standard chemical embeddings as well as a learned, structurally-aware embedding -- SmallSA -- for this task. With this framework, searches on over one billion chemicals execute in less than a second on a single CPU core, five orders of magnitude faster than the brute-force approach. We also demonstrate that SmallSA achieves competitive performance on chemical similarity benchmarks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge