Kjell Gunnar Gundersen
Classifying Dry Eye Disease Patients from Healthy Controls Using Machine Learning and Metabolomics Data
Jun 20, 2024
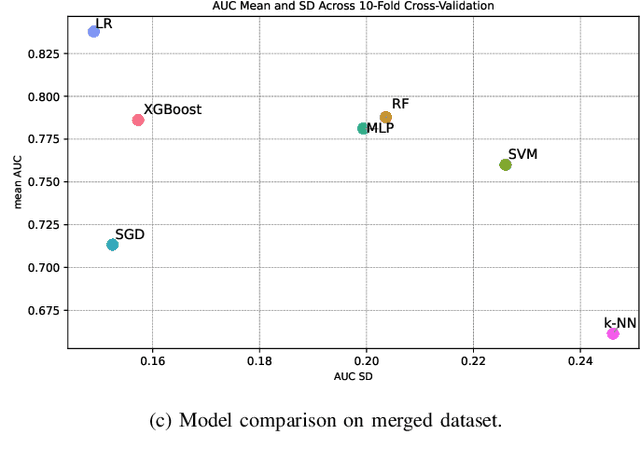
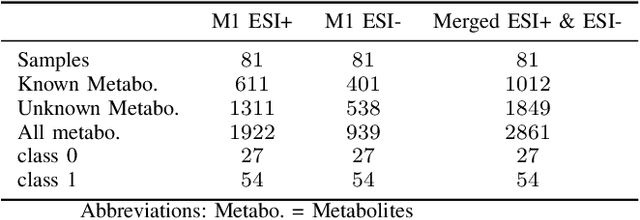

Abstract:Dry eye disease is a common disorder of the ocular surface, leading patients to seek eye care. Clinical signs and symptoms are currently used to diagnose dry eye disease. Metabolomics, a method for analyzing biological systems, has been found helpful in identifying distinct metabolites in patients and in detecting metabolic profiles that may indicate dry eye disease at early stages. In this study, we explored using machine learning and metabolomics information to identify which cataract patients suffered from dry eye disease. As there is no one-size-fits-all machine learning model for metabolomics data, choosing the most suitable model can significantly affect the quality of predictions and subsequent metabolomics analyses. To address this challenge, we conducted a comparative analysis of nine machine learning models on three metabolomics data sets from cataract patients with and without dry eye disease. The models were evaluated and optimized using nested k-fold cross-validation. To assess the performance of these models, we selected a set of suitable evaluation metrics tailored to the data set's challenges. The logistic regression model overall performed the best, achieving the highest area under the curve score of 0.8378, balanced accuracy of 0.735, Matthew's correlation coefficient of 0.5147, an F1-score of 0.8513, and a specificity of 0.5667. Additionally, following the logistic regression, the XGBoost and Random Forest models also demonstrated good performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge