Khalid Raza
Medical Image Generation using Generative Adversarial Networks
May 19, 2020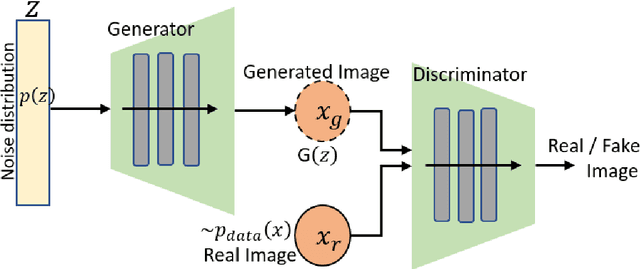
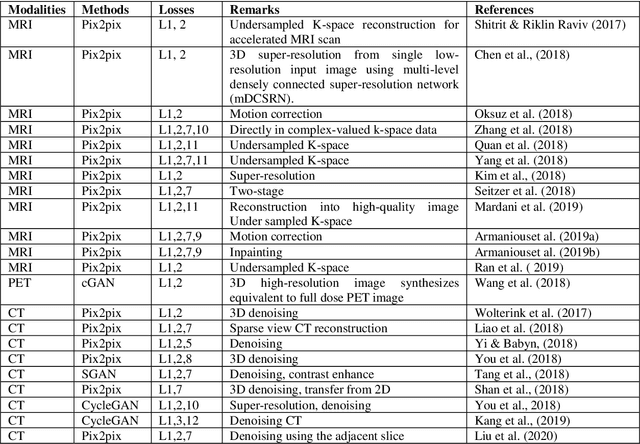
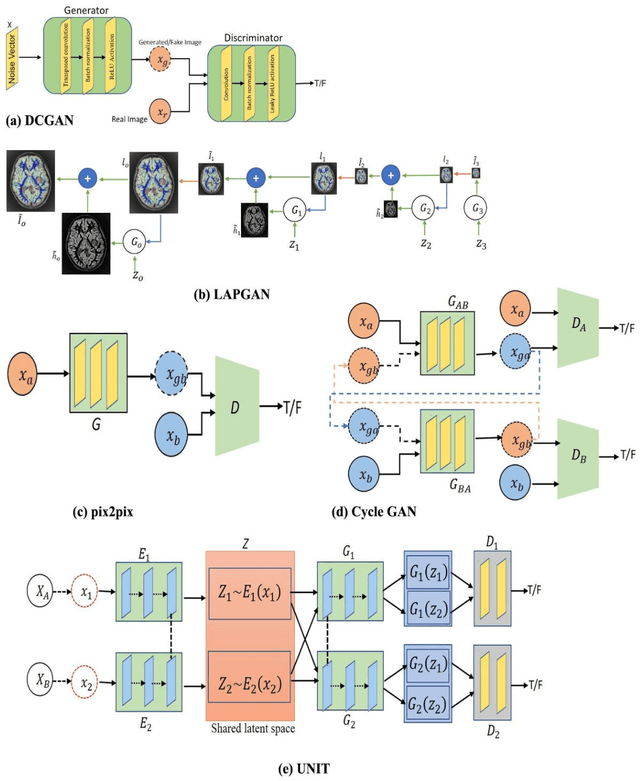
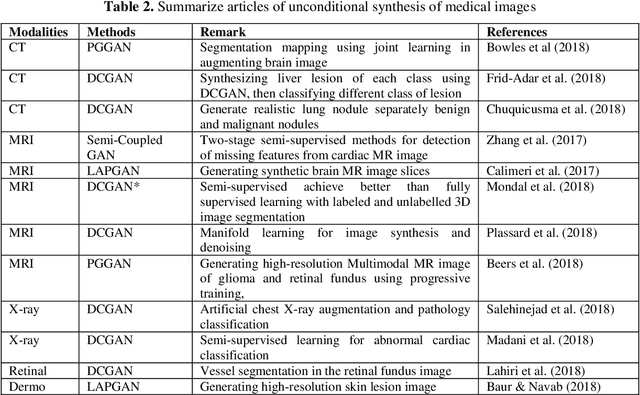
Abstract:Generative adversarial networks (GANs) are unsupervised Deep Learning approach in the computer vision community which has gained significant attention from the last few years in identifying the internal structure of multimodal medical imaging data. The adversarial network simultaneously generates realistic medical images and corresponding annotations, which proven to be useful in many cases such as image augmentation, image registration, medical image generation, image reconstruction, and image-to-image translation. These properties bring the attention of the researcher in the field of medical image analysis and we are witness of rapid adaption in many novel and traditional applications. This chapter provides state-of-the-art progress in GANs-based clinical application in medical image generation, and cross-modality synthesis. The various framework of GANs which gained popularity in the interpretation of medical images, such as Deep Convolutional GAN (DCGAN), Laplacian GAN (LAPGAN), pix2pix, CycleGAN, and unsupervised image-to-image translation model (UNIT), continue to improve their performance by incorporating additional hybrid architecture, has been discussed. Further, some of the recent applications of these frameworks for image reconstruction, and synthesis, and future research directions in the area have been covered.
A Tour of Unsupervised Deep Learning for Medical Image Analysis
Dec 19, 2018
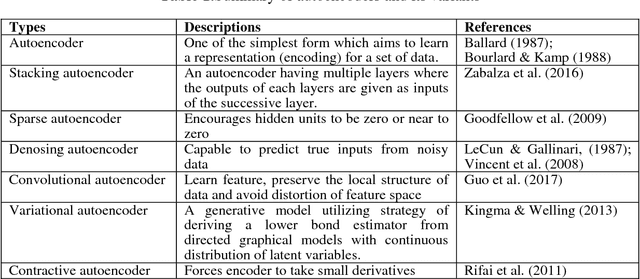
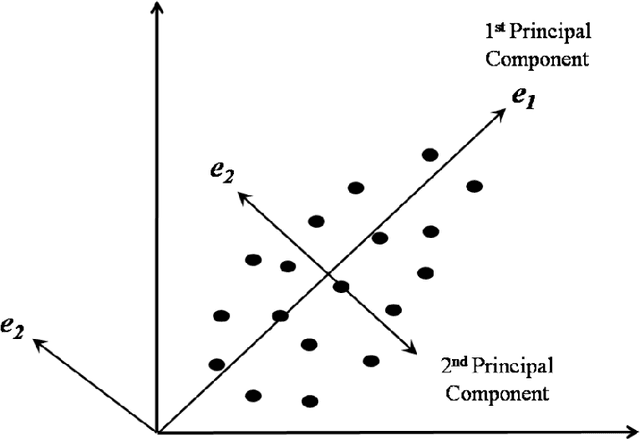
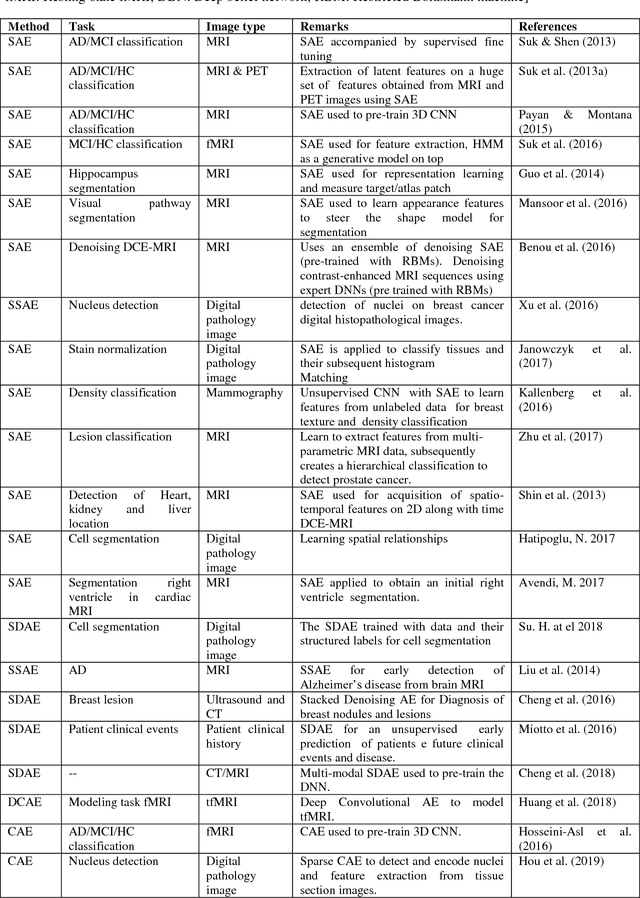
Abstract:Interpretation of medical images for diagnosis and treatment of complex disease from high-dimensional and heterogeneous data remains a key challenge in transforming healthcare. In the last few years, both supervised and unsupervised deep learning achieved promising results in the area of medical imaging and image analysis. Unlike supervised learning which is biased towards how it is being supervised and manual efforts to create class label for the algorithm, unsupervised learning derive insights directly from the data itself, group the data and help to make data driven decisions without any external bias. This review systematically presents various unsupervised models applied to medical image analysis, including autoencoders and its several variants, Restricted Boltzmann machines, Deep belief networks, Deep Boltzmann machine and Generative adversarial network. Future research opportunities and challenges of unsupervised techniques for medical image analysis have also been discussed.
Optimizing Deep Neural Network Architecture: A Tabu Search Based Approach
Aug 17, 2018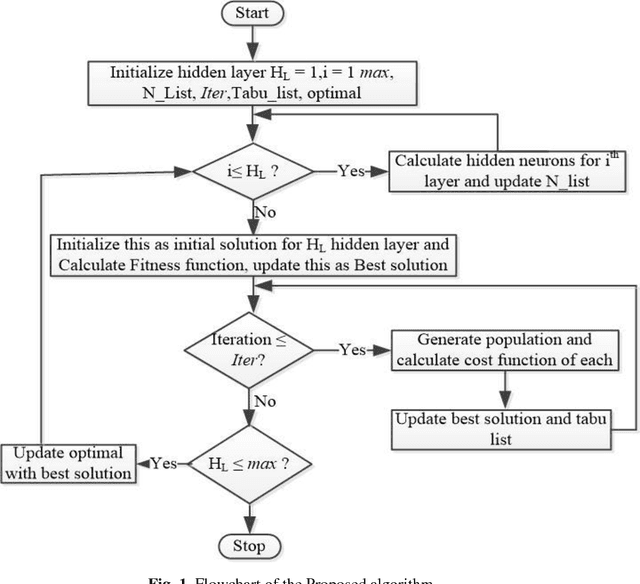
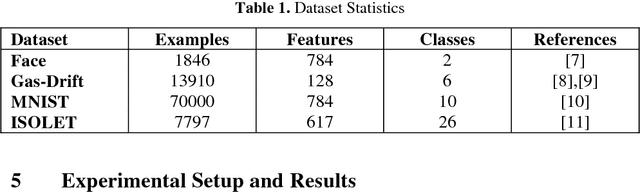
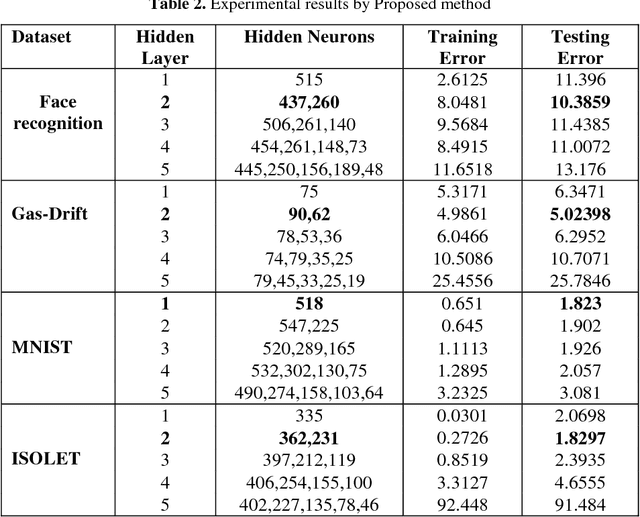
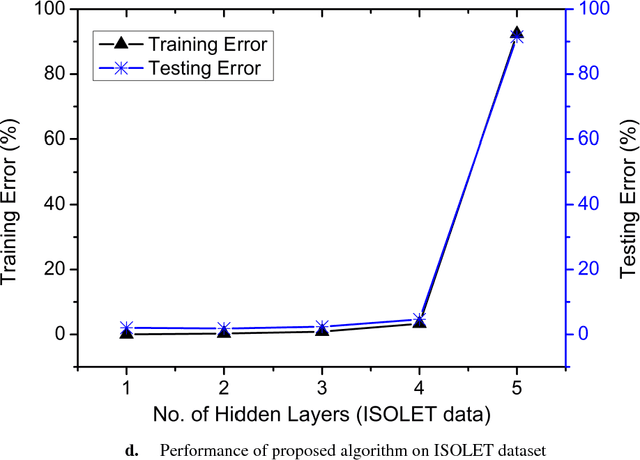
Abstract:The performance of Feedforward neural network (FNN) fully de-pends upon the selection of architecture and training algorithm. FNN architecture can be tweaked using several parameters, such as the number of hidden layers, number of hidden neurons at each hidden layer and number of connections between layers. There may be exponential combinations for these architectural attributes which may be unmanageable manually, so it requires an algorithm which can automatically design an optimal architecture with high generalization ability. Numerous optimization algorithms have been utilized for FNN architecture determination. This paper proposes a new methodology which can work on the estimation of hidden layers and their respective neurons for FNN. This work combines the advantages of Tabu search (TS) and Gradient descent with momentum backpropagation (GDM) training algorithm to demonstrate how Tabu search can automatically select the best architecture from the populated architectures based on minimum testing error criteria. The proposed approach has been tested on four classification benchmark dataset of different size.
Recurrent Neural Network Based Hybrid Model of Gene Regulatory Network
Nov 13, 2015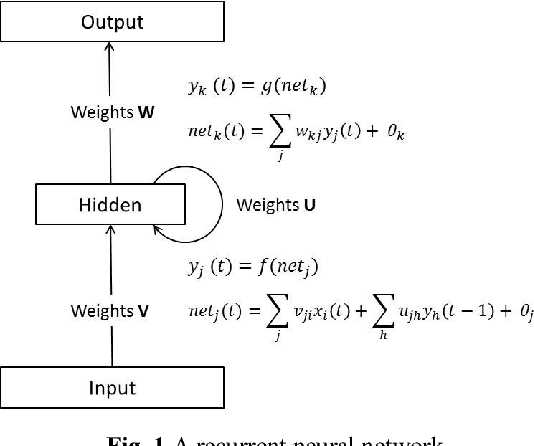
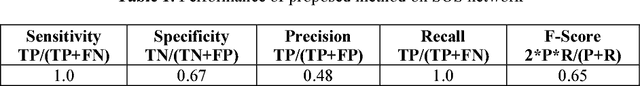
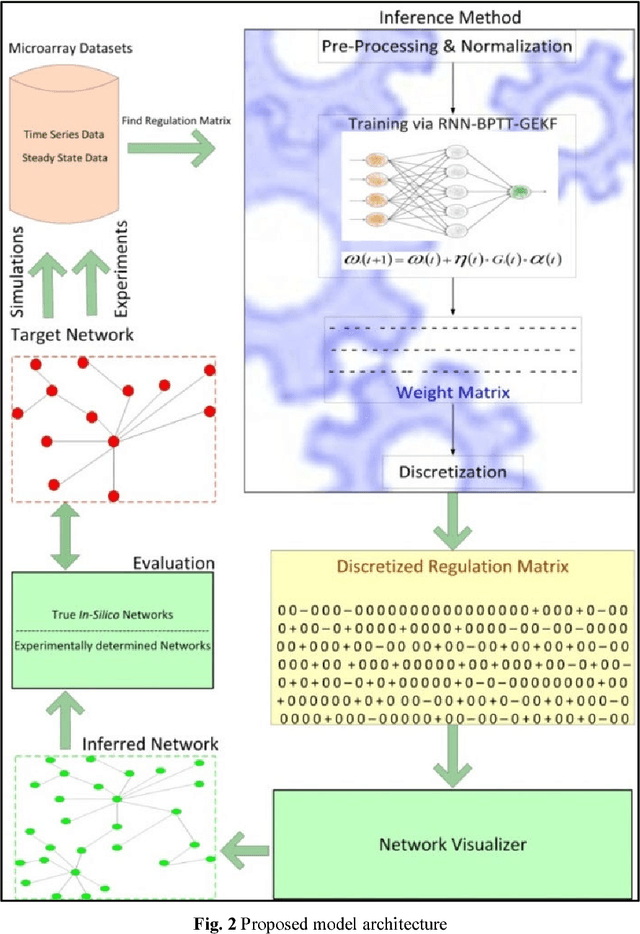
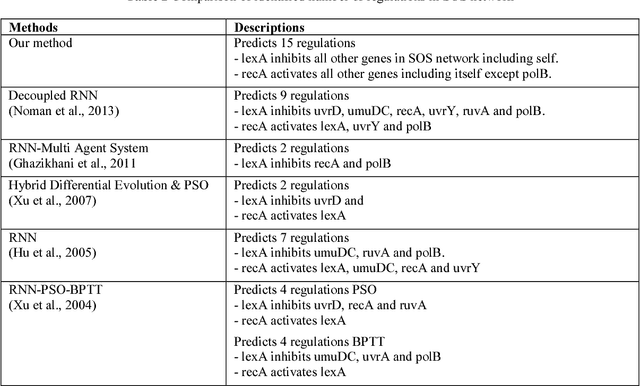
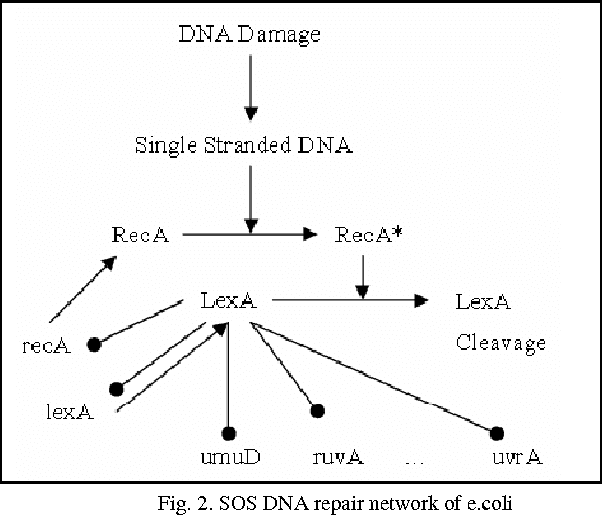
Abstract:Systems biology is an emerging interdisciplinary area of research that focuses on study of complex interactions in a biological system, such as gene regulatory networks. The discovery of gene regulatory networks leads to a wide range of applications, such as pathways related to a disease that can unveil in what way the disease acts and provide novel tentative drug targets. In addition, the development of biological models from discovered networks or pathways can help to predict the responses to disease and can be much useful for the novel drug development and treatments. The inference of regulatory networks from biological data is still in its infancy stage. This paper proposes a recurrent neural network (RNN) based gene regulatory network (GRN) model hybridized with generalized extended Kalman filter for weight update in backpropagation through time training algorithm. The RNN is a complex neural network that gives a better settlement between the biological closeness and mathematical flexibility to model GRN. The RNN is able to capture complex, non-linear and dynamic relationship among variables. Gene expression data are inherently noisy and Kalman filter performs well for estimation even in noisy data. Hence, non-linear version of Kalman filter, i.e., generalized extended Kalman filter has been applied for weight update during network training. The developed model has been applied on DNA SOS repair network, IRMA network, and two synthetic networks from DREAM Challenge. We compared our results with other state-of-the-art techniques that show superiority of our model. Further, 5% Gaussian noise has been added in the dataset and result of the proposed model shows negligible effect of noise on the results.
* 18 pages, 9 figures and 4 tables
Analysis of Microarray Data using Artificial Intelligence Based Techniques
Jul 10, 2015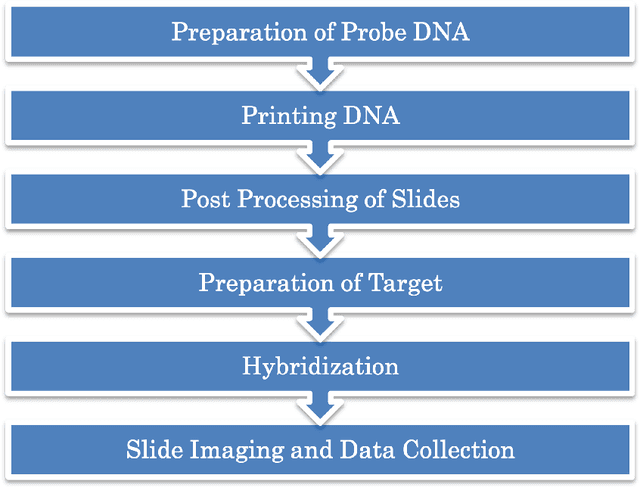
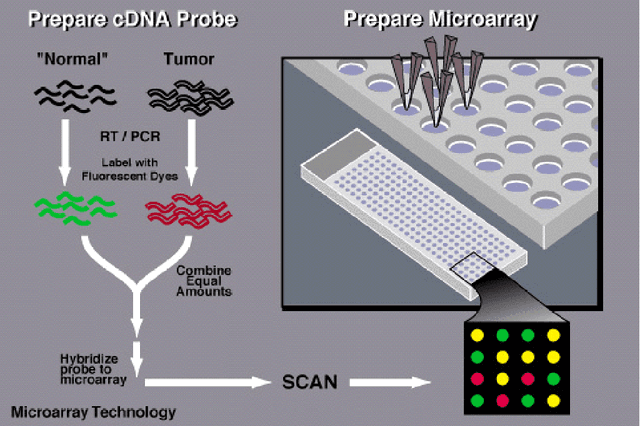
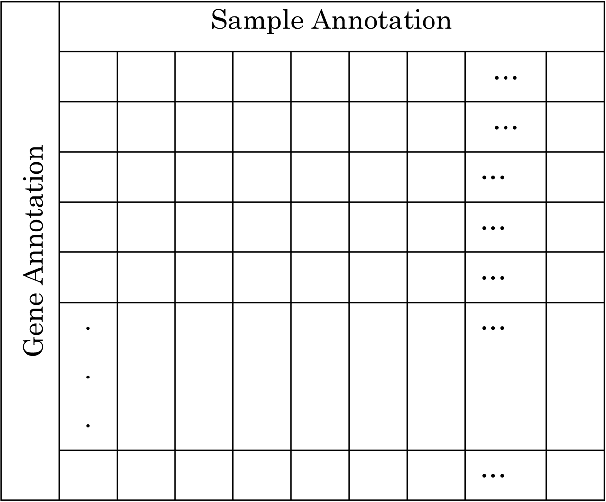
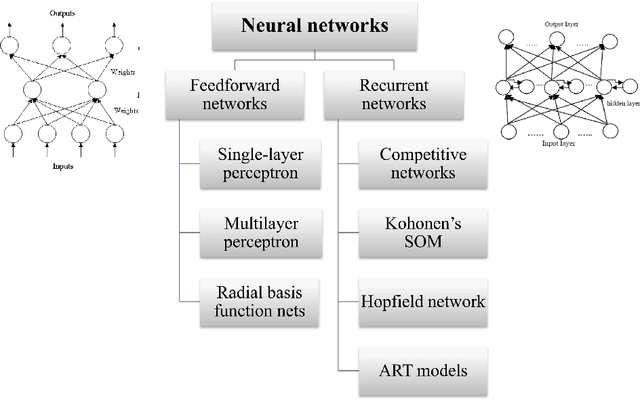
Abstract:Microarray is one of the essential technologies used by the biologist to measure genome-wide expression levels of genes in a particular organism under some particular conditions or stimuli. As microarrays technologies have become more prevalent, the challenges of analyzing these data for getting better insight about biological processes have essentially increased. Due to availability of artificial intelligence based sophisticated computational techniques, such as artificial neural networks, fuzzy logic, genetic algorithms, and many other nature-inspired algorithms, it is possible to analyse microarray gene expression data in more better way. Here, we reviewed artificial intelligence based techniques for the analysis of microarray gene expression data. Further, challenges in the field and future work direction have also been suggested.
* 32 pages, 4 figures
Formal Concept Analysis for Knowledge Discovery from Biological Data
Jun 01, 2015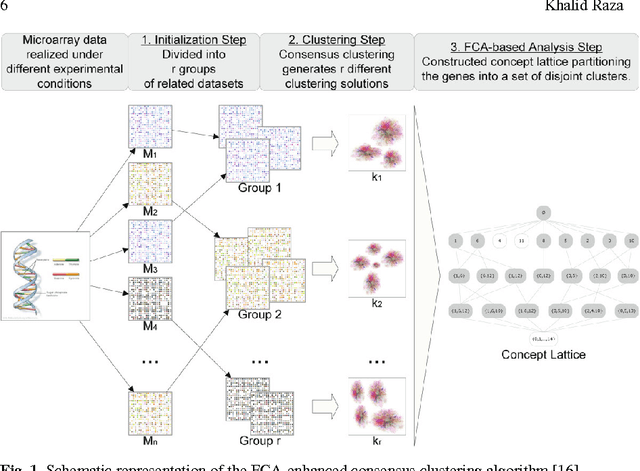
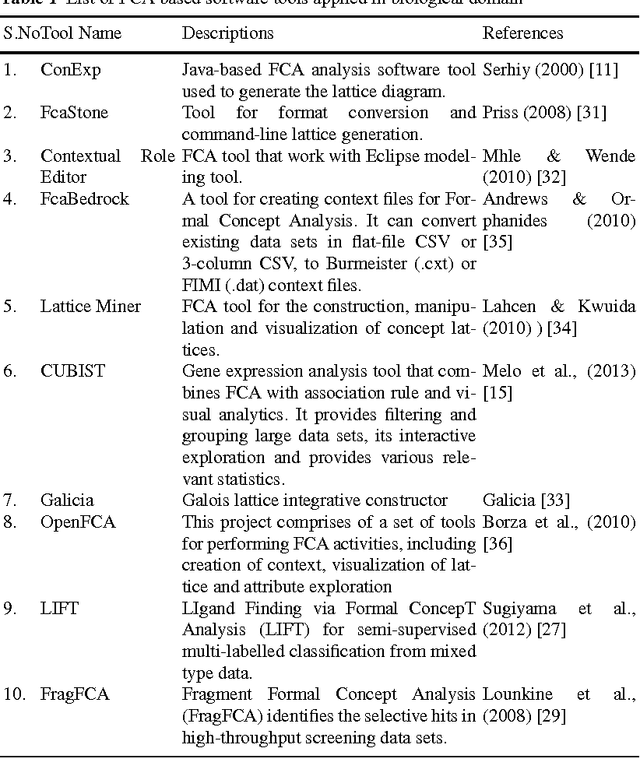
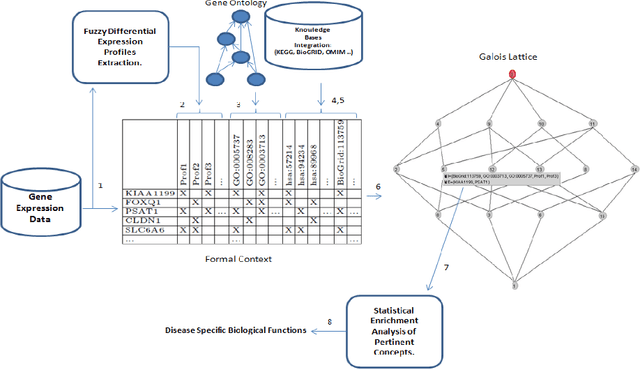
Abstract:Due to rapid advancement in high-throughput techniques, such as microarrays and next generation sequencing technologies, biological data are increasing exponentially. The current challenge in computational biology and bioinformatics research is how to analyze these huge raw biological data to extract biologically meaningful knowledge. This review paper presents the applications of formal concept analysis for the analysis and knowledge discovery from biological data, including gene expression discretization, gene co-expression mining, gene expression clustering, finding genes in gene regulatory networks, enzyme/protein classifications, binding site classifications, and so on. It also presents a list of FCA-based software tools applied in biological domain and covers the challenges faced so far.
Ant Colony Optimization for Inferring Key Gene Interactions
Jun 06, 2014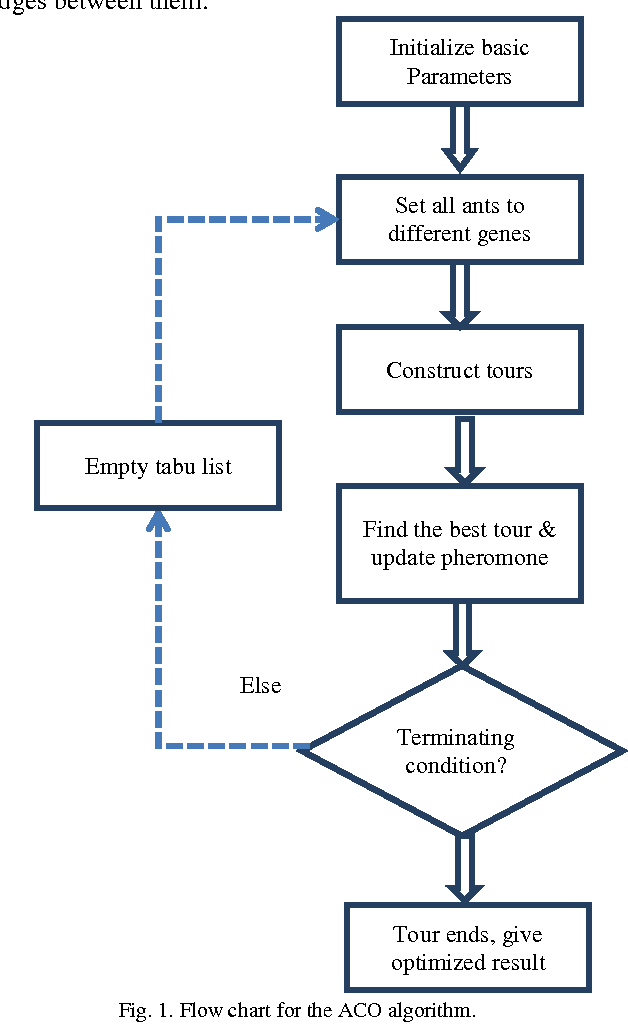
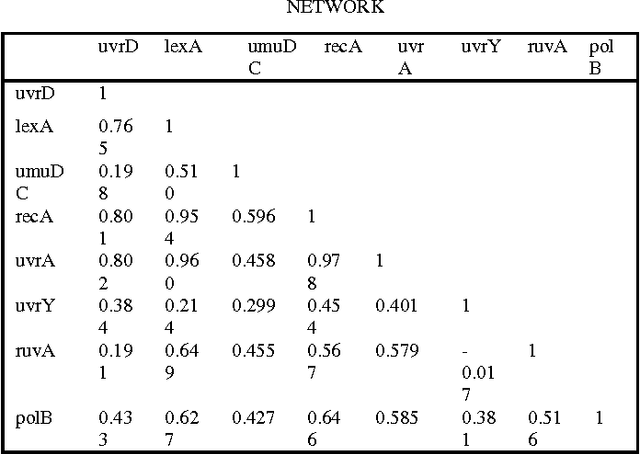
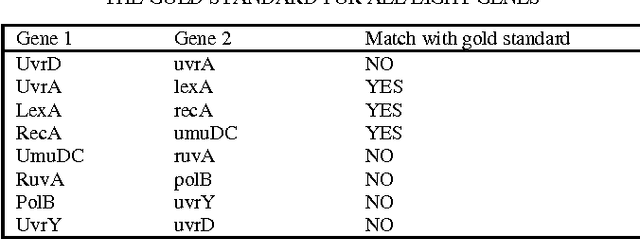
Abstract:Inferring gene interaction network from gene expression data is an important task in systems biology research. The gene interaction network, especially key interactions, plays an important role in identifying biomarkers for disease that further helps in drug design. Ant colony optimization is an optimization algorithm based on natural evolution and has been used in many optimization problems. In this paper, we applied ant colony optimization algorithm for inferring the key gene interactions from gene expression data. The algorithm has been tested on two different kinds of benchmark datasets and observed that it successfully identify some key gene interactions.
* 8 pages, 2 figures and 4 tables
A Comprehensive Evaluation of Machine Learning Techniques for Cancer Class Prediction Based on Microarray Data
Jul 26, 2013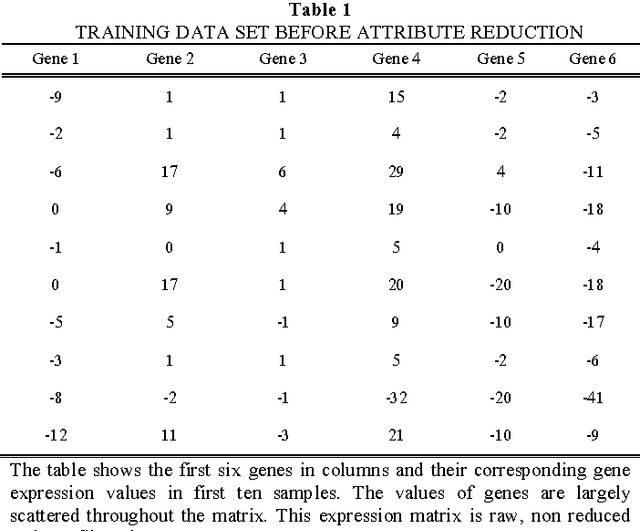
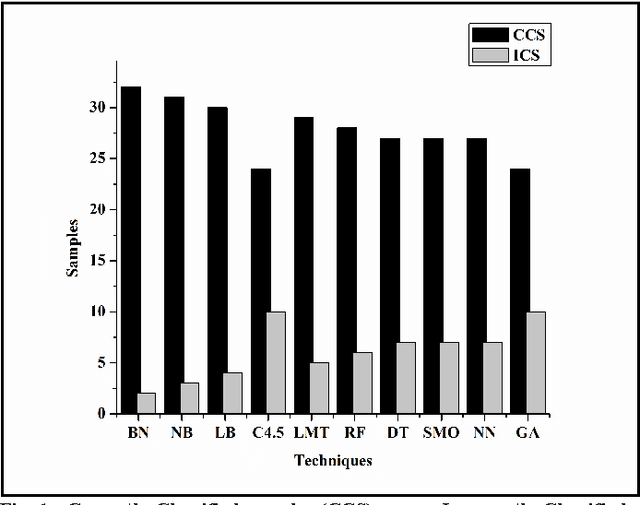
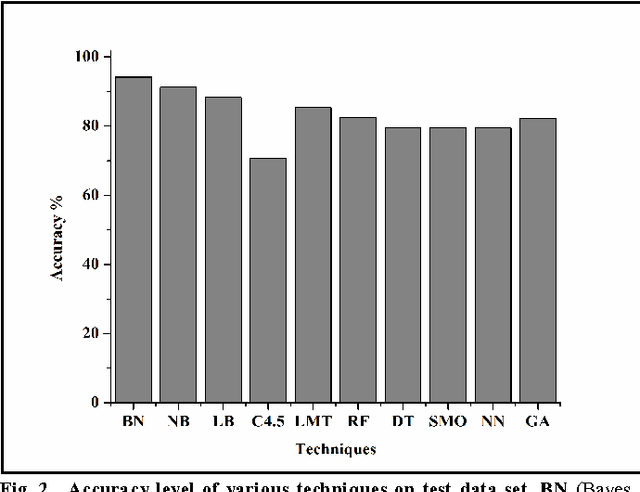
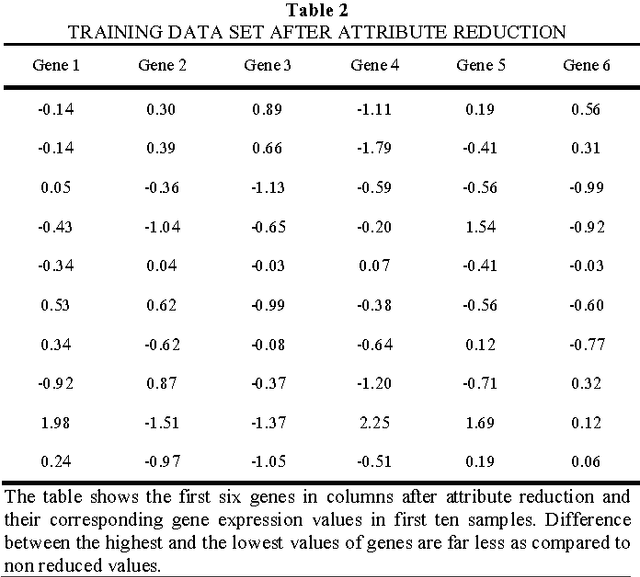
Abstract:Prostate cancer is among the most common cancer in males and its heterogeneity is well known. Its early detection helps making therapeutic decision. There is no standard technique or procedure yet which is full-proof in predicting cancer class. The genomic level changes can be detected in gene expression data and those changes may serve as standard model for any random cancer data for class prediction. Various techniques were implied on prostate cancer data set in order to accurately predict cancer class including machine learning techniques. Huge number of attributes and few number of sample in microarray data leads to poor machine learning, therefore the most challenging part is attribute reduction or non significant gene reduction. In this work we have compared several machine learning techniques for their accuracy in predicting the cancer class. Machine learning is effective when number of attributes (genes) are larger than the number of samples which is rarely possible with gene expression data. Attribute reduction or gene filtering is absolutely required in order to make the data more meaningful as most of the genes do not participate in tumor development and are irrelevant for cancer prediction. Here we have applied combination of statistical techniques such as inter-quartile range and t-test, which has been effective in filtering significant genes and minimizing noise from data. Further we have done a comprehensive evaluation of ten state-of-the-art machine learning techniques for their accuracy in class prediction of prostate cancer. Out of these techniques, Bayes Network out performed with an accuracy of 94.11% followed by Navie Bayes with an accuracy of 91.17%. To cross validate our results, we modified our training dataset in six different way and found that average sensitivity, specificity, precision and accuracy of Bayes Network is highest among all other techniques used.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge