Atif N Hasan
A Comprehensive Evaluation of Machine Learning Techniques for Cancer Class Prediction Based on Microarray Data
Jul 26, 2013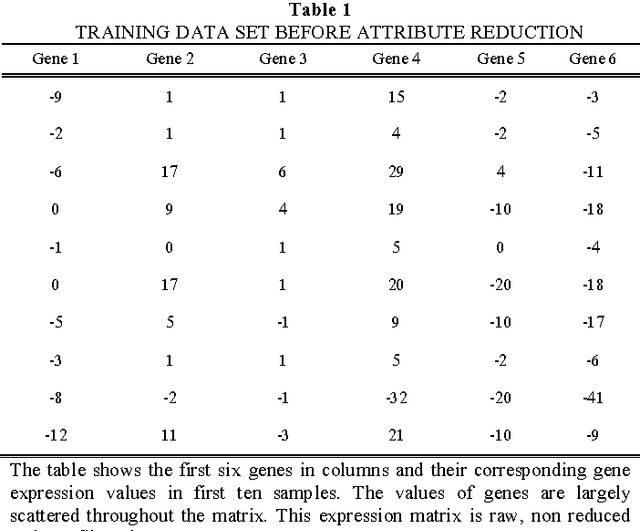
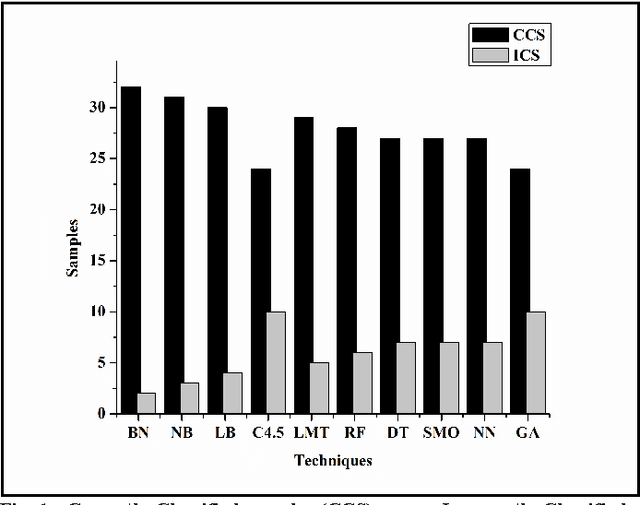
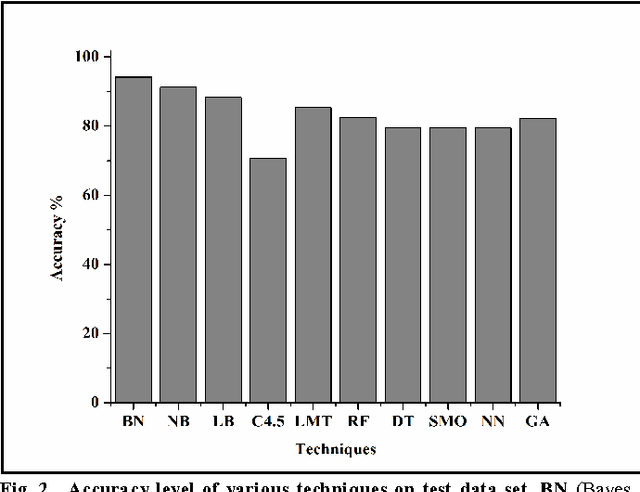
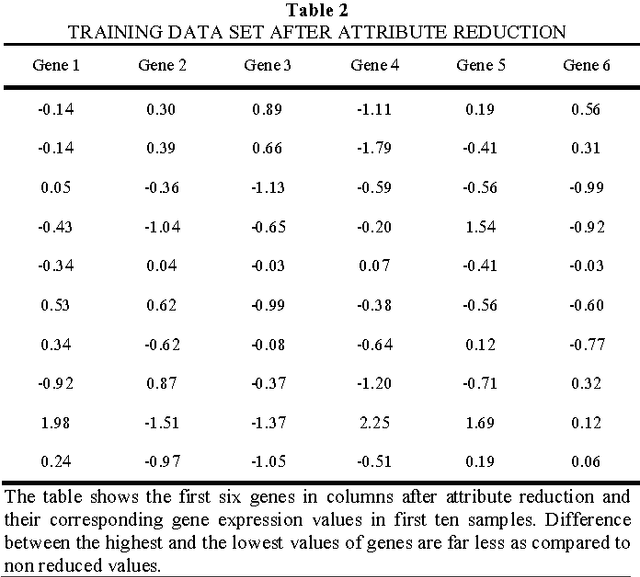
Abstract:Prostate cancer is among the most common cancer in males and its heterogeneity is well known. Its early detection helps making therapeutic decision. There is no standard technique or procedure yet which is full-proof in predicting cancer class. The genomic level changes can be detected in gene expression data and those changes may serve as standard model for any random cancer data for class prediction. Various techniques were implied on prostate cancer data set in order to accurately predict cancer class including machine learning techniques. Huge number of attributes and few number of sample in microarray data leads to poor machine learning, therefore the most challenging part is attribute reduction or non significant gene reduction. In this work we have compared several machine learning techniques for their accuracy in predicting the cancer class. Machine learning is effective when number of attributes (genes) are larger than the number of samples which is rarely possible with gene expression data. Attribute reduction or gene filtering is absolutely required in order to make the data more meaningful as most of the genes do not participate in tumor development and are irrelevant for cancer prediction. Here we have applied combination of statistical techniques such as inter-quartile range and t-test, which has been effective in filtering significant genes and minimizing noise from data. Further we have done a comprehensive evaluation of ten state-of-the-art machine learning techniques for their accuracy in class prediction of prostate cancer. Out of these techniques, Bayes Network out performed with an accuracy of 94.11% followed by Navie Bayes with an accuracy of 91.17%. To cross validate our results, we modified our training dataset in six different way and found that average sensitivity, specificity, precision and accuracy of Bayes Network is highest among all other techniques used.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge