Karishma Thakrar
StAyaL | Multilingual Style Transfer
Jan 20, 2025



Abstract:Stylistic text generation plays a vital role in enhancing communication by reflecting the nuances of individual expression. This paper presents a novel approach for generating text in a specific speaker's style across different languages. We show that by leveraging only 100 lines of text, an individuals unique style can be captured as a high-dimensional embedding, which can be used for both text generation and stylistic translation. This methodology breaks down the language barrier by transferring the style of a speaker between languages. The paper is structured into three main phases: augmenting the speaker's data with stylistically consistent external sources, separating style from content using machine learning and deep learning techniques, and generating an abstract style profile by mean pooling the learned embeddings. The proposed approach is shown to be topic-agnostic, with test accuracy and F1 scores of 74.9\% and 0.75, respectively. The results demonstrate the potential of the style profile for multilingual communication, paving the way for further applications in personalized content generation and cross-linguistic stylistic transfer.
Enhancing Talent Employment Insights Through Feature Extraction with LLM Finetuning
Jan 13, 2025Abstract:This paper explores the application of large language models (LLMs) to extract nuanced and complex job features from unstructured job postings. Using a dataset of 1.2 million job postings provided by AdeptID, we developed a robust pipeline to identify and classify variables such as remote work availability, remuneration structures, educational requirements, and work experience preferences. Our methodology combines semantic chunking, retrieval-augmented generation (RAG), and fine-tuning DistilBERT models to overcome the limitations of traditional parsing tools. By leveraging these techniques, we achieved significant improvements in identifying variables often mislabeled or overlooked, such as non-salary-based compensation and inferred remote work categories. We present a comprehensive evaluation of our fine-tuned models and analyze their strengths, limitations, and potential for scaling. This work highlights the promise of LLMs in labor market analytics, providing a foundation for more accurate and actionable insights into job data.
AlgoRxplorers | Precision in Mutation -- Enhancing Drug Design with Advanced Protein Stability Prediction Tools
Jan 13, 2025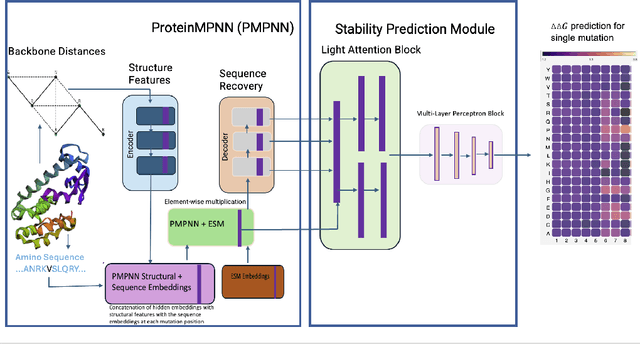
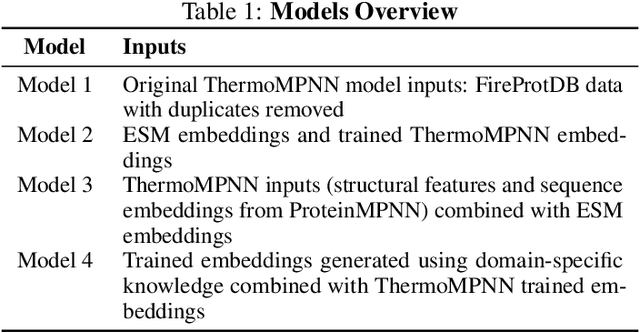
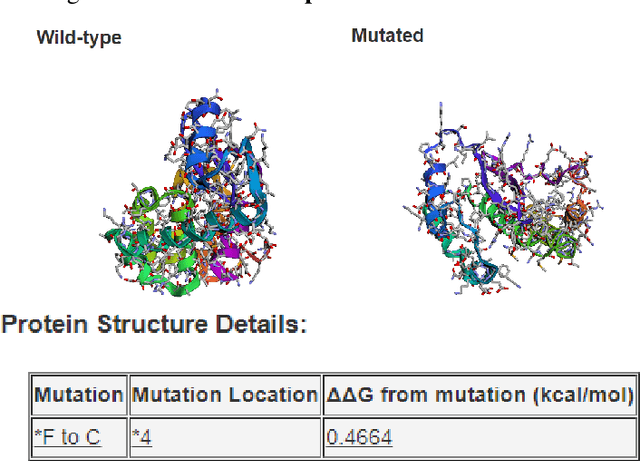
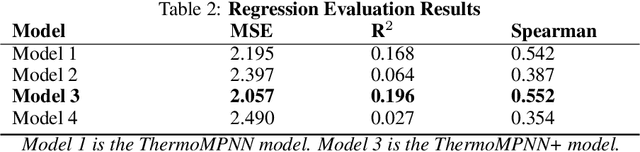
Abstract:Predicting the impact of single-point amino acid mutations on protein stability is essential for understanding disease mechanisms and advancing drug development. Protein stability, quantified by changes in Gibbs free energy ($\Delta\Delta G$), is influenced by these mutations. However, the scarcity of data and the complexity of model interpretation pose challenges in accurately predicting stability changes. This study proposes the application of deep neural networks, leveraging transfer learning and fusing complementary information from different models, to create a feature-rich representation of the protein stability landscape. We developed four models, with our third model, ThermoMPNN+, demonstrating the best performance in predicting $\Delta\Delta G$ values. This approach, which integrates diverse feature sets and embeddings through latent transfusion techniques, aims to refine $\Delta\Delta G$ predictions and contribute to a deeper understanding of protein dynamics, potentially leading to advancements in disease research and drug discovery.
DynaGRAG: Improving Language Understanding and Generation through Dynamic Subgraph Representation in Graph Retrieval-Augmented Generation
Dec 24, 2024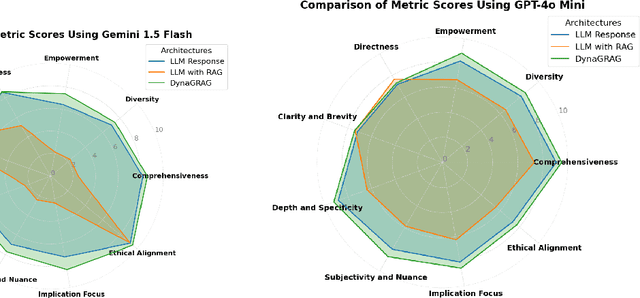
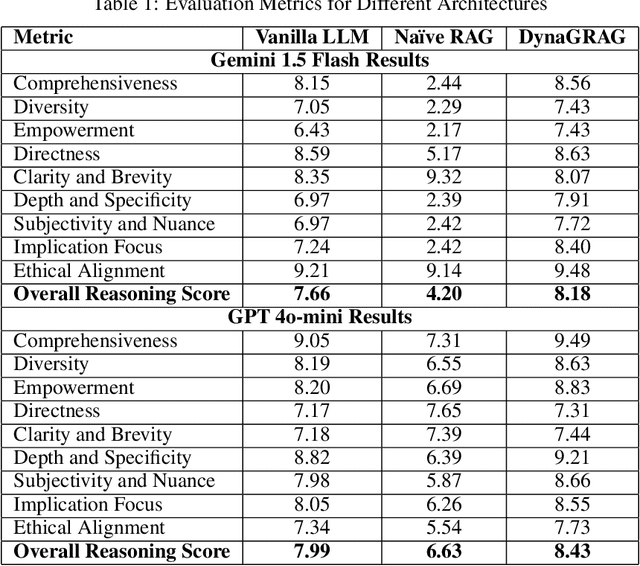
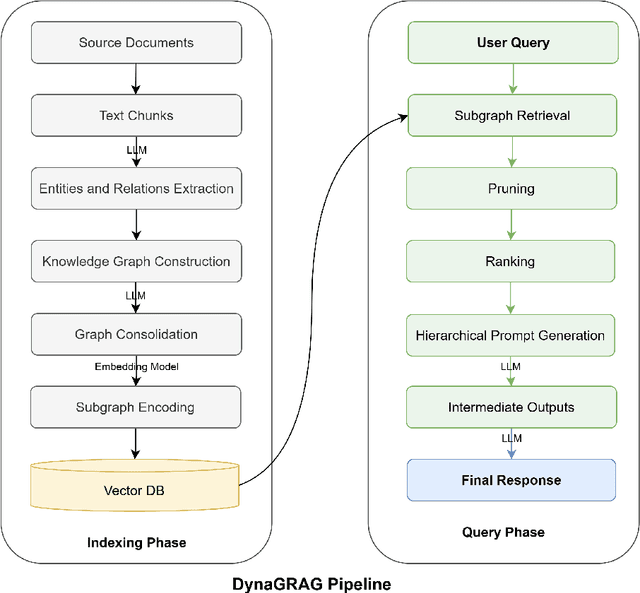
Abstract:Graph Retrieval-Augmented Generation (GRAG or Graph RAG) architectures aim to enhance language understanding and generation by leveraging external knowledge. However, effectively capturing and integrating the rich semantic information present in textual and structured data remains a challenge. To address this, a novel GRAG framework is proposed to focus on enhancing subgraph representation and diversity within the knowledge graph. By improving graph density, capturing entity and relation information more effectively, and dynamically prioritizing relevant and diverse subgraphs, the proposed approach enables a more comprehensive understanding of the underlying semantic structure. This is achieved through a combination of de-duplication processes, two-step mean pooling of embeddings, query-aware retrieval considering unique nodes, and a Dynamic Similarity-Aware BFS (DSA-BFS) traversal algorithm. Integrating Graph Convolutional Networks (GCNs) and Large Language Models (LLMs) through hard prompting further enhances the learning of rich node and edge representations while preserving the hierarchical subgraph structure. Experimental results on multiple benchmark datasets demonstrate the effectiveness of the proposed GRAG framework, showcasing the significance of enhanced subgraph representation and diversity for improved language understanding and generation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge