Jules Grégory
ONCOPILOT: A Promptable CT Foundation Model For Solid Tumor Evaluation
Oct 10, 2024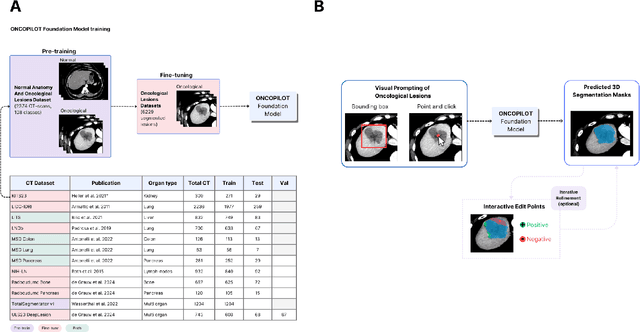
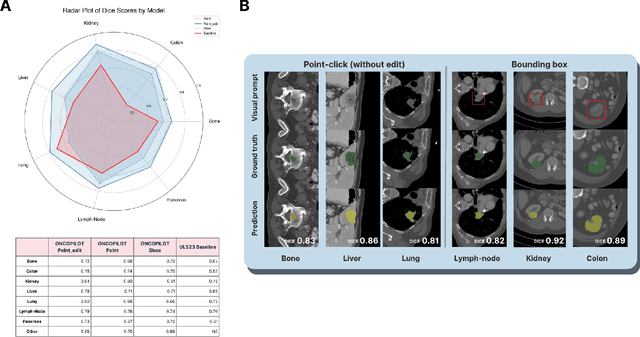
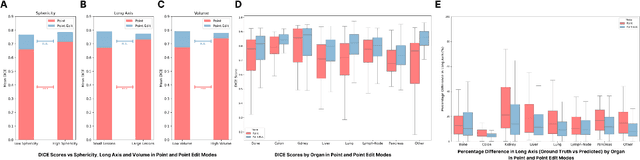
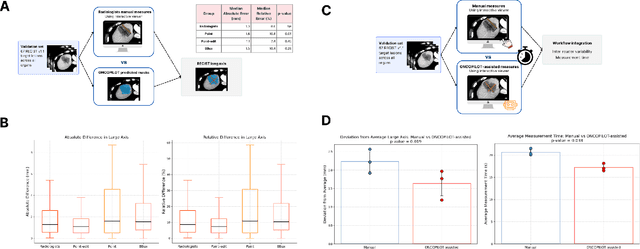
Abstract:Carcinogenesis is a proteiform phenomenon, with tumors emerging in various locations and displaying complex, diverse shapes. At the crucial intersection of research and clinical practice, it demands precise and flexible assessment. However, current biomarkers, such as RECIST 1.1's long and short axis measurements, fall short of capturing this complexity, offering an approximate estimate of tumor burden and a simplistic representation of a more intricate process. Additionally, existing supervised AI models face challenges in addressing the variability in tumor presentations, limiting their clinical utility. These limitations arise from the scarcity of annotations and the models' focus on narrowly defined tasks. To address these challenges, we developed ONCOPILOT, an interactive radiological foundation model trained on approximately 7,500 CT scans covering the whole body, from both normal anatomy and a wide range of oncological cases. ONCOPILOT performs 3D tumor segmentation using visual prompts like point-click and bounding boxes, outperforming state-of-the-art models (e.g., nnUnet) and achieving radiologist-level accuracy in RECIST 1.1 measurements. The key advantage of this foundation model is its ability to surpass state-of-the-art performance while keeping the radiologist in the loop, a capability that previous models could not achieve. When radiologists interactively refine the segmentations, accuracy improves further. ONCOPILOT also accelerates measurement processes and reduces inter-reader variability, facilitating volumetric analysis and unlocking new biomarkers for deeper insights. This AI assistant is expected to enhance the precision of RECIST 1.1 measurements, unlock the potential of volumetric biomarkers, and improve patient stratification and clinical care, while seamlessly integrating into the radiological workflow.
Primary liver cancer classification from routine tumour biopsy using weakly supervised deep learning
Apr 07, 2024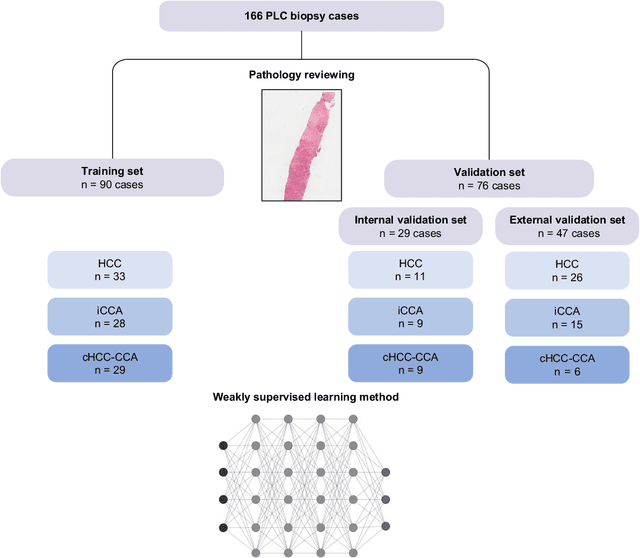
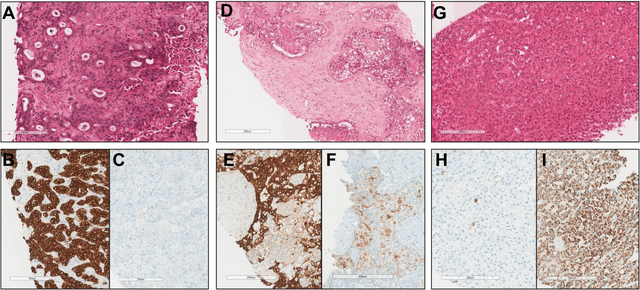
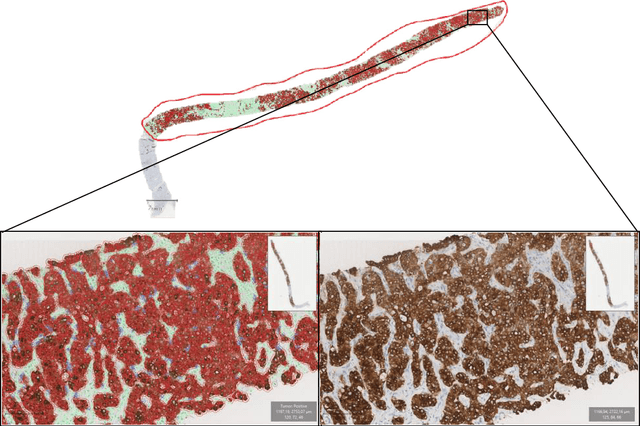
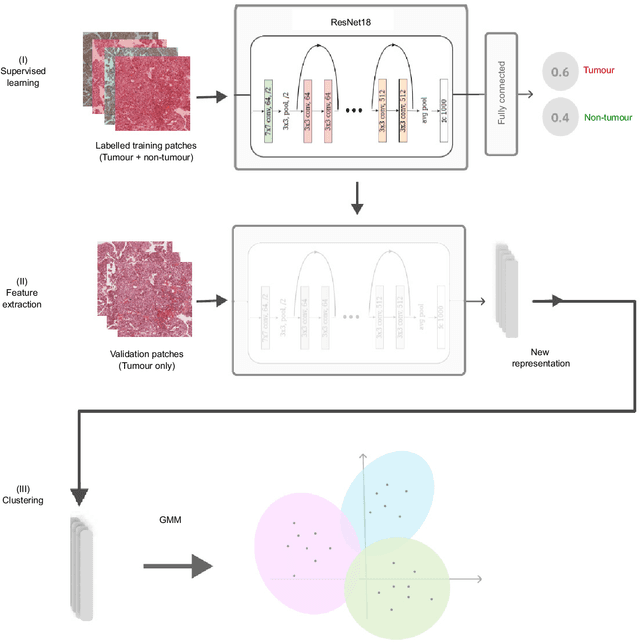
Abstract:The diagnosis of primary liver cancers (PLCs) can be challenging, especially on biopsies and for combined hepatocellular-cholangiocarcinoma (cHCC-CCA). We automatically classified PLCs on routine-stained biopsies using a weakly supervised learning method. Weak tumour/non-tumour annotations served as labels for training a Resnet18 neural network, and the network's last convolutional layer was used to extract new tumour tile features. Without knowledge of the precise labels of the malignancies, we then applied an unsupervised clustering algorithm. Our model identified specific features of hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma (iCCA). Despite no specific features of cHCC-CCA being recognized, the identification of HCC and iCCA tiles within a slide could facilitate the diagnosis of primary liver cancers, particularly cHCC-CCA. Method and results: 166 PLC biopsies were divided into training, internal and external validation sets: 90, 29 and 47 samples. Two liver pathologists reviewed each whole-slide hematein eosin saffron (HES)-stained image (WSI). After annotating the tumour/non-tumour areas, 256x256 pixel tiles were extracted from the WSIs and used to train a ResNet18. The network was used to extract new tile features. An unsupervised clustering algorithm was then applied to the new tile features. In a two-cluster model, Clusters 0 and 1 contained mainly HCC and iCCA histological features. The diagnostic agreement between the pathological diagnosis and the model predictions in the internal and external validation sets was 100% (11/11) and 96% (25/26) for HCC and 78% (7/9) and 87% (13/15) for iCCA, respectively. For cHCC-CCA, we observed a highly variable proportion of tiles from each cluster (Cluster 0: 5-97%; Cluster 1: 2-94%).
* https://www.sciencedirect.com/science/article/pii/S2589555924000090
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge