Jintao Bai
Super-resolution reconstruction of cytoskeleton image based on A-net deep learning network
Dec 17, 2021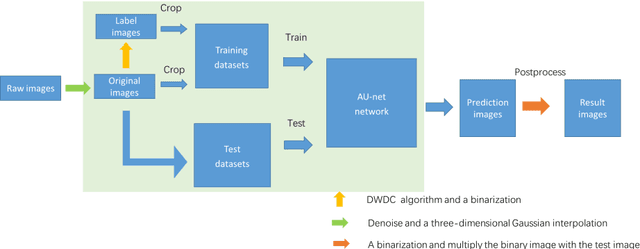
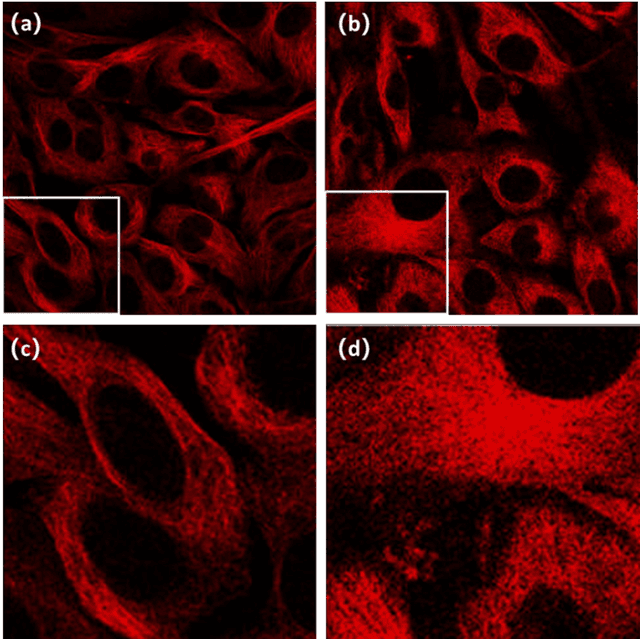
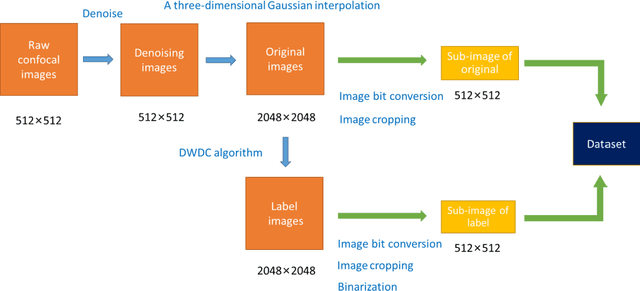
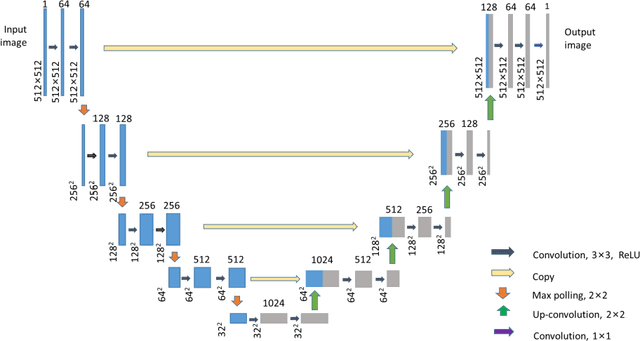
Abstract:To date, live-cell imaging at the nanometer scale remains challenging. Even though super-resolution microscopy methods have enabled visualization of subcellular structures below the optical resolution limit, the spatial resolution is still far from enough for the structural reconstruction of biomolecules in vivo (i.e. ~24 nm thickness of microtubule fiber). In this study, we proposed an A-net network and showed that the resolution of cytoskeleton images captured by a confocal microscope can be significantly improved by combining the A-net deep learning network with the DWDC algorithm based on degradation model. Utilizing the DWDC algorithm to construct new datasets and taking advantage of A-net neural network's features (i.e., considerably fewer layers), we successfully removed the noise and flocculent structures, which originally interfere with the cellular structure in the raw image, and improved the spatial resolution by 10 times using relatively small dataset. We, therefore, conclude that the proposed algorithm that combines A-net neural network with the DWDC method is a suitable and universal approach for exacting structural details of biomolecules, cells and organs from low-resolution images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge