Jingyu Lee
Liver Segmentation in Abdominal CT Images via Auto-Context Neural Network and Self-Supervised Contour Attention
Feb 14, 2020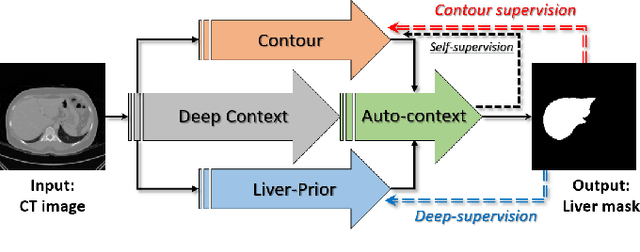
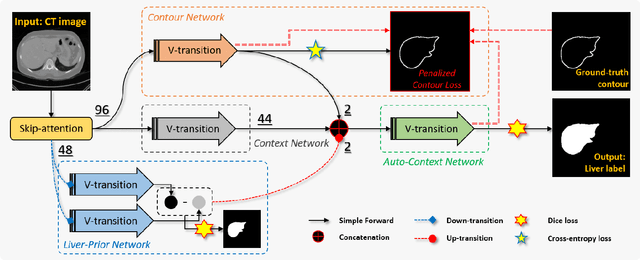
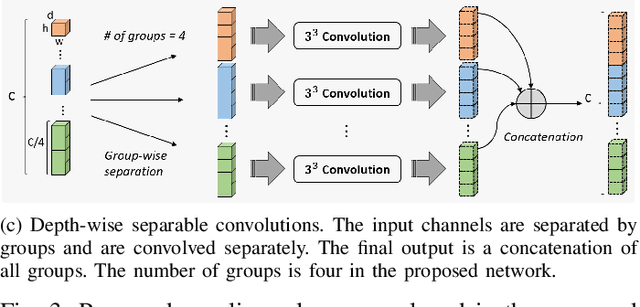
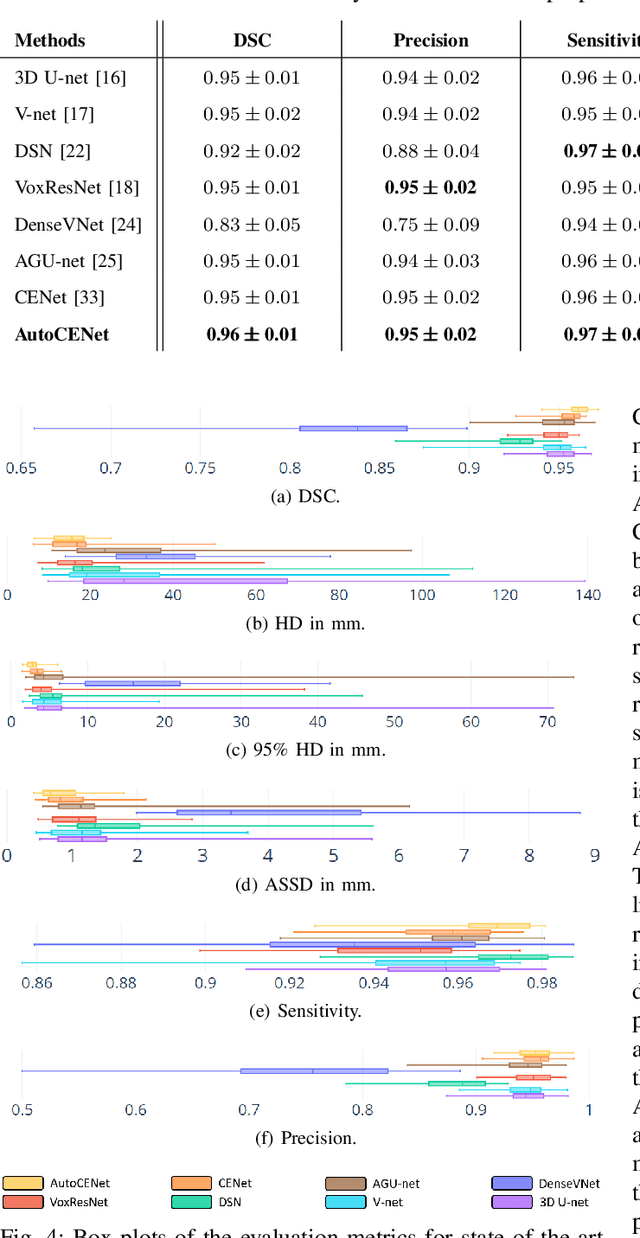
Abstract:Accurate image segmentation of the liver is a challenging problem owing to its large shape variability and unclear boundaries. Although the applications of fully convolutional neural networks (CNNs) have shown groundbreaking results, limited studies have focused on the performance of generalization. In this study, we introduce a CNN for liver segmentation on abdominal computed tomography (CT) images that shows high generalization performance and accuracy. To improve the generalization performance, we initially propose an auto-context algorithm in a single CNN. The proposed auto-context neural network exploits an effective high-level residual estimation to obtain the shape prior. Identical dual paths are effectively trained to represent mutual complementary features for an accurate posterior analysis of a liver. Further, we extend our network by employing a self-supervised contour scheme. We trained sparse contour features by penalizing the ground-truth contour to focus more contour attentions on the failures. The experimental results show that the proposed network results in better accuracy when compared to the state-of-the-art networks by reducing 10.31% of the Hausdorff distance. We used 180 abdominal CT images for training and validation. Two-fold cross-validation is presented for a comparison with the state-of-the-art neural networks. Novel multiple N-fold cross-validations are conducted to verify the performance of generalization. The proposed network showed the best generalization performance among the networks. Additionally, we present a series of ablation experiments that comprehensively support the importance of the underlying concepts.
Pose-Aware Instance Segmentation Framework from Cone Beam CT Images for Tooth Segmentation
Feb 06, 2020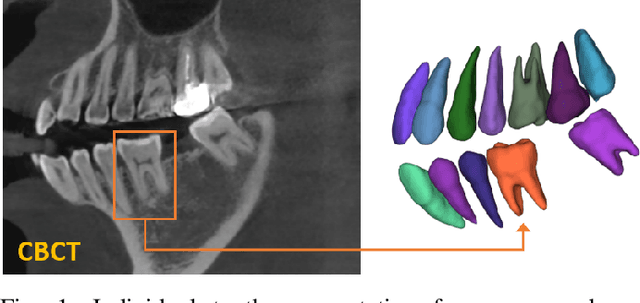
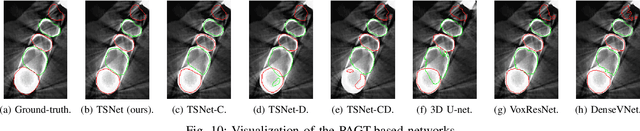
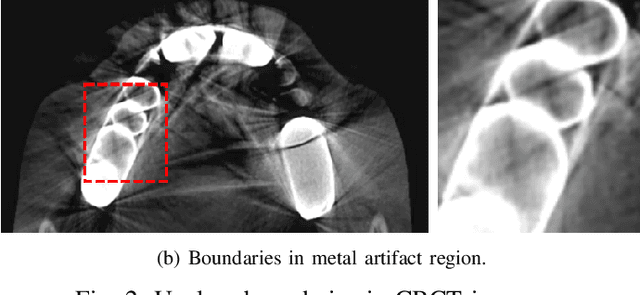
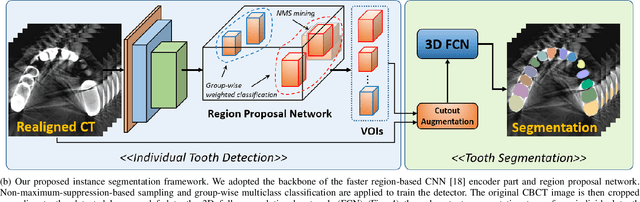
Abstract:Individual tooth segmentation from cone beam computed tomography (CBCT) images is an essential prerequisite for an anatomical understanding of orthodontic structures in several applications, such as tooth reformation planning and implant guide simulations. However, the presence of severe metal artifacts in CBCT images hinders the accurate segmentation of each individual tooth. In this study, we propose a neural network for pixel-wise labeling to exploit an instance segmentation framework that is robust to metal artifacts. Our method comprises of three steps: 1) image cropping and realignment by pose regressions, 2) metal-robust individual tooth detection, and 3) segmentation. We first extract the alignment information of the patient by pose regression neural networks to attain a volume-of-interest (VOI) region and realign the input image, which reduces the inter-overlapping area between tooth bounding boxes. Then, individual tooth regions are localized within a VOI realigned image using a convolutional detector. We improved the accuracy of the detector by employing non-maximum suppression and multiclass classification metrics in the region proposal network. Finally, we apply a convolutional neural network (CNN) to perform individual tooth segmentation by converting the pixel-wise labeling task to a distance regression task. Metal-intensive image augmentation is also employed for a robust segmentation of metal artifacts. The result shows that our proposed method outperforms other state-of-the-art methods, especially for teeth with metal artifacts. The primary significance of the proposed method is two-fold: 1) an introduction of pose-aware VOI realignment followed by a robust tooth detection and 2) a metal-robust CNN framework for accurate tooth segmentation.
Automatic Registration between Cone-Beam CT and Scanned Surface via Deep-Pose Regression Neural Networks and Clustered Similarities
Jul 29, 2019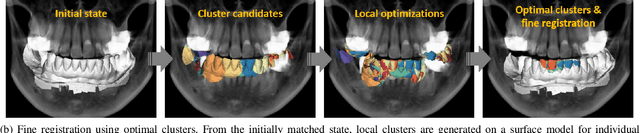
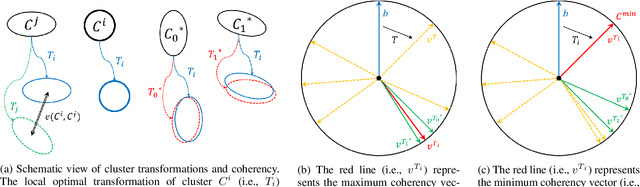
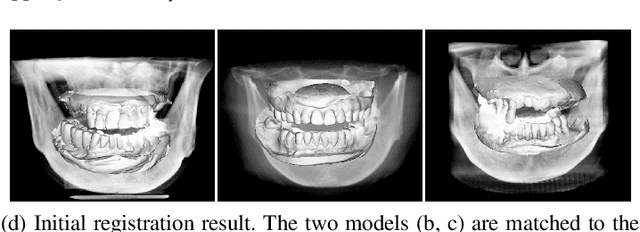
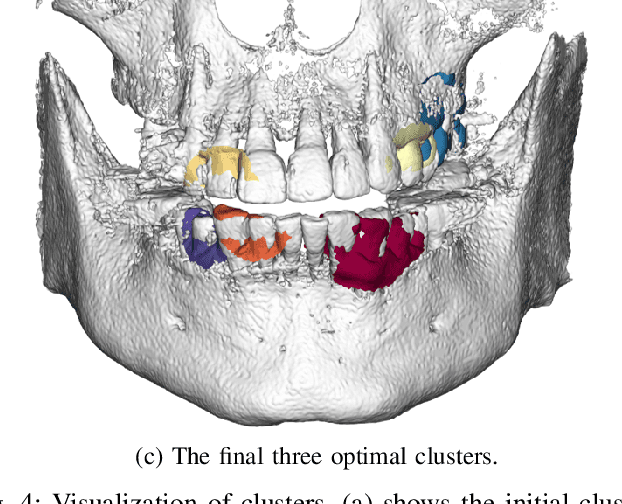
Abstract:Computerized registration between maxillofacial cone-beam computed tomography (CT) images and a scanned dental model is an essential prerequisite in surgical planning for dental implants or orthognathic surgery. We propose a novel method that performs fully automatic registration between a cone-beam CT image and an optically scanned model. To build a robust and automatic initial registration method, our method applies deep-pose regression neural networks in a reduced domain (i.e., 2-dimensional image). Subsequently, fine registration is performed via optimal clusters. Majority voting system achieves globally optimal transformations while each cluster attempts to optimize local transformation parameters. The coherency of clusters determines their candidacy for the optimal cluster set. The outlying regions in the iso-surface are effectively removed based on the consensus among the optimal clusters. The accuracy of registration was evaluated by the Euclidean distance of 10 landmarks on a scanned model which were annotated by the experts in the field. The experiments show that the proposed method's registration accuracy, measured in landmark distance, outperforms other existing methods by 30.77% to 70%. In addition to achieving high accuracy, our proposed method requires neither human-interactions nor priors (e.g., iso-surface extraction). The main significance of our study is twofold: 1) the employment of light-weighted neural networks which indicates the applicability of neural network in extracting pose cues that can be easily obtained and 2) the introduction of an optimal cluster-based registration method that can avoid metal artifacts during the matching procedures.
Deeply Self-Supervising Edge-to-Contour Neural Network Applied to Liver Segmentation
Aug 06, 2018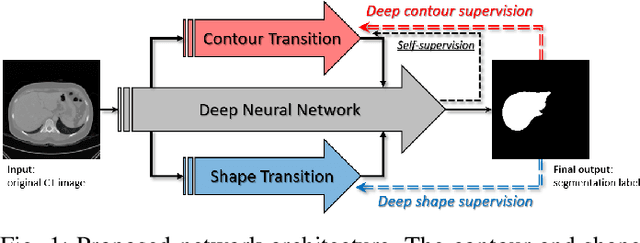
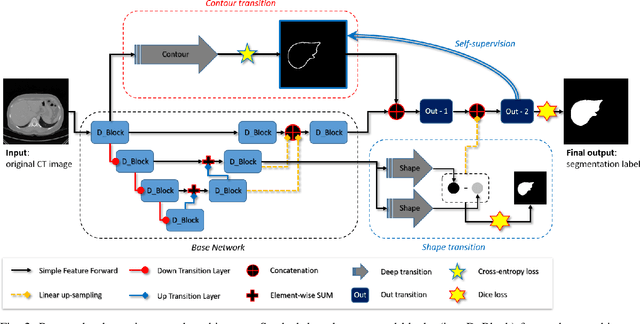
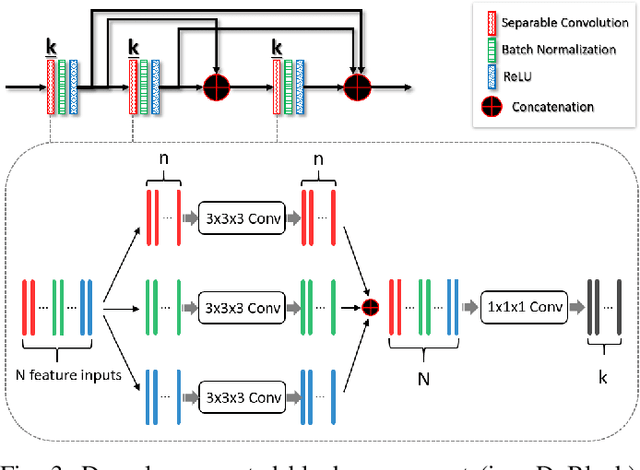
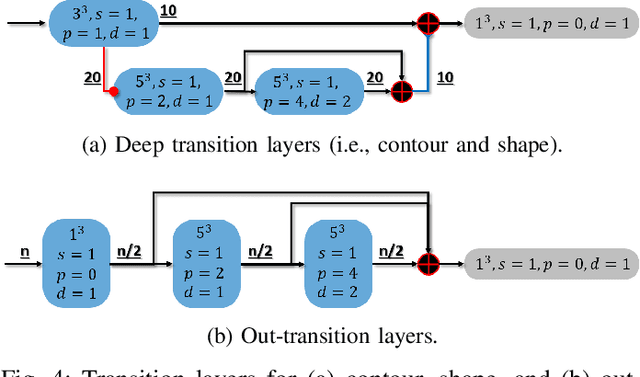
Abstract:Accurate segmentation of liver is still a challenging problem due to its large shape variability and unclear boundaries. The purpose of this paper is to propose a neural network based liver segmentation algorithm and evaluate its performance on abdominal CT images. First, we develop a fully convolutional network (FCN) for volumetric image segmentation problem. To guide a neural network to accurately delineate the target liver object, we apply self-supervising scheme with respect to edge and contour responses. The deeply supervising method is also applied to our low-level features for further combining discriminative features in the higher feature dimensions. We used 160 abdominal CT images for training and validation. Quantitative evaluation of our proposed network is presented with 8-fold cross-validation. The result showed that our method successfully segmented liver more accurately than any other state-of-the-art methods without expanding or deepening the neural network. The proposed approach can be easily extended to other imaging protocols (e.g., MRI) or other target organ segmentation problems without any modifications of the framework.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge