Jer Hayes
Evaluation of the Synthetic Electronic Health Records
Oct 16, 2022


Abstract:Generative models have been found effective for data synthesis due to their ability to capture complex underlying data distributions. The quality of generated data from these models is commonly evaluated by visual inspection for image datasets or downstream analytical tasks for tabular datasets. These evaluation methods neither measure the implicit data distribution nor consider the data privacy issues, and it remains an open question of how to compare and rank different generative models. Medical data can be sensitive, so it is of great importance to draw privacy concerns of patients while maintaining the data utility of the synthetic dataset. Beyond the utility evaluation, this work outlines two metrics called Similarity and Uniqueness for sample-wise assessment of synthetic datasets. We demonstrate the proposed notions with several state-of-the-art generative models to synthesise Cystic Fibrosis (CF) patients' electronic health records (EHRs), observing that the proposed metrics are suitable for synthetic data evaluation and generative model comparison.
Synthesising Electronic Health Records: Cystic Fibrosis Patient Group
Jan 14, 2022



Abstract:Class imbalance can often degrade predictive performance of supervised learning algorithms. Balanced classes can be obtained by oversampling exact copies, with noise, or interpolation between nearest neighbours (as in traditional SMOTE methods). Oversampling tabular data using augmentation, as is typical in computer vision tasks, can be achieved with deep generative models. Deep generative models are effective data synthesisers due to their ability to capture complex underlying distributions. Synthetic data in healthcare can enhance interoperability between healthcare providers by ensuring patient privacy. Equipped with large synthetic datasets which do well to represent small patient groups, machine learning in healthcare can address the current challenges of bias and generalisability. This paper evaluates synthetic data generators ability to synthesise patient electronic health records. We test the utility of synthetic data for patient outcome classification, observing increased predictive performance when augmenting imbalanced datasets with synthetic data.
Network Generation with Differential Privacy
Nov 17, 2021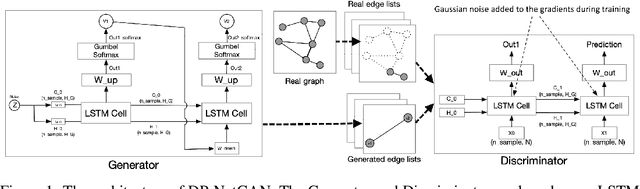
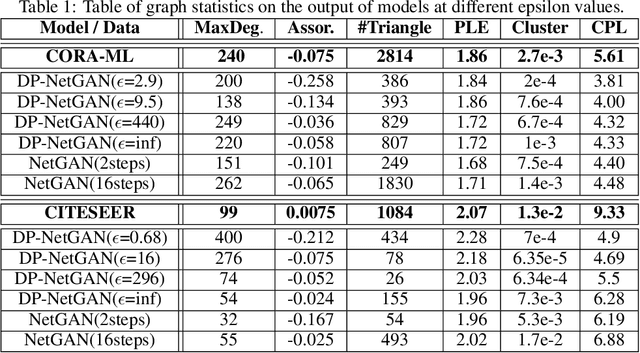
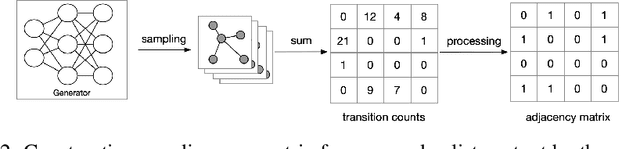
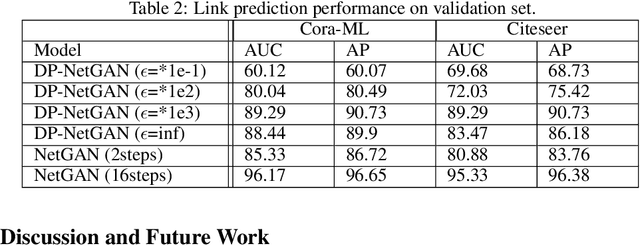
Abstract:We consider the problem of generating private synthetic versions of real-world graphs containing private information while maintaining the utility of generated graphs. Differential privacy is a gold standard for data privacy, and the introduction of the differentially private stochastic gradient descent (DP-SGD) algorithm has facilitated the training of private neural models in a number of domains. Recent advances in graph generation via deep generative networks have produced several high performing models. We evaluate and compare state-of-the-art models including adjacency matrix based models and edge based models, and show a practical implementation that favours the edge-list approach utilizing the Gaussian noise mechanism when evaluated on commonly used graph datasets. Based on our findings, we propose a generative model that can reproduce the properties of real-world networks while maintaining edge-differential privacy. The proposed model is based on a stochastic neural network that generates discrete edge-list samples and is trained using the Wasserstein GAN objective with the DP-SGD optimizer. Being the first approach to combine these beneficial properties, our model contributes to further research on graph data privacy.
A Graph-based Imputation Method for Sparse Medical Records
Nov 17, 2021



Abstract:Electronic Medical Records (EHR) are extremely sparse. Only a small proportion of events (symptoms, diagnoses, and treatments) are observed in the lifetime of an individual. The high degree of missingness of EHR can be attributed to a large number of factors, including device failure, privacy concerns, or other unexpected reasons. Unfortunately, many traditional imputation methods are not well suited for highly sparse data and scale poorly to high dimensional datasets. In this paper, we propose a graph-based imputation method that is both robust to sparsity and to unreliable unmeasured events. Our approach compares favourably to several standard and state-of-the-art imputation methods in terms of performance and runtime. Moreover, results indicate that the model learns to embed different event types in a clinically meaningful way. Our work can facilitate the diagnosis of novel diseases based on the clinical history of past events, with the potential to increase our understanding of the landscape of comorbidities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge