Jens Haueisen
Scatter-based common spatial patterns -- a unified spatial filtering framework
Mar 07, 2023Abstract:The common spatial pattern (CSP) approach is known as one of the most popular spatial filtering techniques for EEG classification in motor imagery (MI) based brain-computer interfaces (BCIs). However, it still suffers some drawbacks such as sensitivity to noise, non-stationarity, and limitation to binary classification.Therefore, we propose a novel spatial filtering framework called scaCSP based on the scatter matrices of spatial covariances of EEG signals, which works generally in both binary and multi-class problems whereas CSP can be cast into our framework as a special case when only the range space of the between-class scatter matrix is used in binary cases.We further propose subspace enhanced scaCSP algorithms which easily permit incorporating more discriminative information contained in other range spaces and null spaces of the between-class and within-class scatter matrices in two scenarios: a nullspace components reduction scenario and an additional spatial filter learning scenario.The proposed algorithms are evaluated on two data sets including 4 MI tasks. The classification performance is compared against state-of-the-art competing algorithms: CSP, Tikhonov regularized CSP (TRCSP), stationary CSP (sCSP) and stationary TRCSP (sTRCSP) in the binary problems whilst multi-class extensions of CSP based on pair-wise and one-versus-rest techniques in the multi-class problems. The results show that the proposed framework outperforms all the competing algorithms in terms of average classification accuracy and computational efficiency in both binary and multi-class problems.The proposed scsCSP works as a unified framework for general multi-class problems and is promising for improving the performance of MI-BCIs.
Online functional connectivity analysis of large all-to-all networks
Mar 06, 2023Abstract:The analysis of EEG/MEG functional connectivity has become an important tool in neural research. Especially the high time resolution of EEG/MEG enables important insight into the functioning of the human brain. To date, functional connectivity is commonly estimated offline, i.e., after the conclusion of the experiment. However, online computation of functional connectivity has the potential to enable unique experimental paradigms. For example, changes of functional connectivity due to learning processes could be tracked in real time and the experiment be adjusted based on these observations. Furthermore, the connectivity estimates can be used for neurofeedback applications or the instantaneous inspection of measurement results. In this study, we present the implementation and evaluation of online sensor and source space functional connectivity estimation in the open-source software MNE Scan. Online capable implementations of several functional connectivity metrics were established in the Connectivity library within MNE-CPP and made available as a plugin in MNE Scan. Online capability was achieved by enforcing multithreading and high efficiency for all computations, so that repeated computations were avoided wherever possible, which allows for a major speed-up in the case of overlapping intervalls. We present comprehensive performance evaluations of these implementations proving the online capability for the computation of large all-to-all functional connectivity networks. As a proof of principle, we demonstrate the feasibility of online functional connectivity estimation in the evaluation of somatosensory evoked brain activity.
Robust Multi-dimensional Model Order Estimation Using LineAr Regression of Global Eigenvalues (LaRGE)
Oct 06, 2021

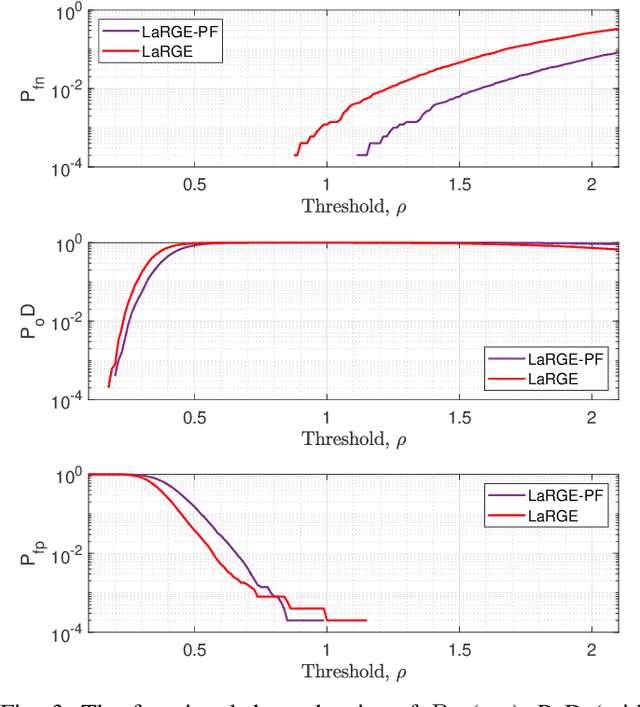

Abstract:The efficient estimation of an approximate model order is very important for real applications with multi-dimensional data if the observed low-rank data is corrupted by additive noise. In this paper, we present a novel robust method for model order estimation of noise-corrupted multi-dimensional low-rank data based on the LineAr Regression of Global Eigenvalues (LaRGE). The LaRGE method uses the multi-linear singular values obtained from the HOSVD of the measurement tensor to construct global eigenvalues. In contrast to the Modified Exponential Test (EFT) that also exploits the approximate exponential profile of the noise eigenvalues, LaRGE does not require the calculation of the probability of false alarm. Moreover, LaRGE achieves a significantly improved performance in comparison with popular state-of-the-art methods. It is well suited for the analysis of biomedical data. The excellent performance of the LaRGE method is illustrated via simulations and results obtained from EEG recordings.
The iterative reweighted Mixed-Norm Estimate for spatio-temporal MEG/EEG source reconstruction
Jul 28, 2016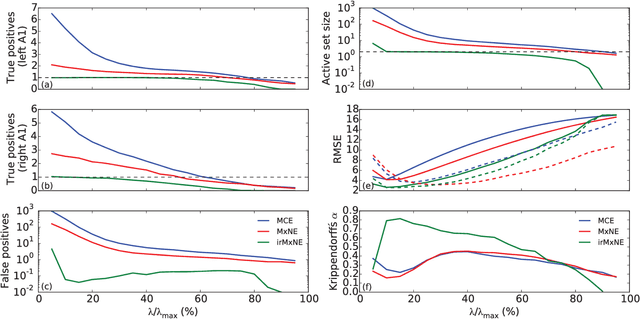
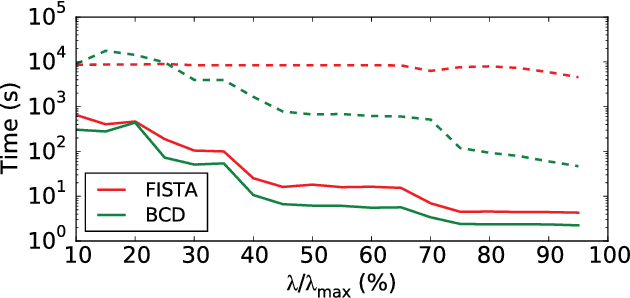
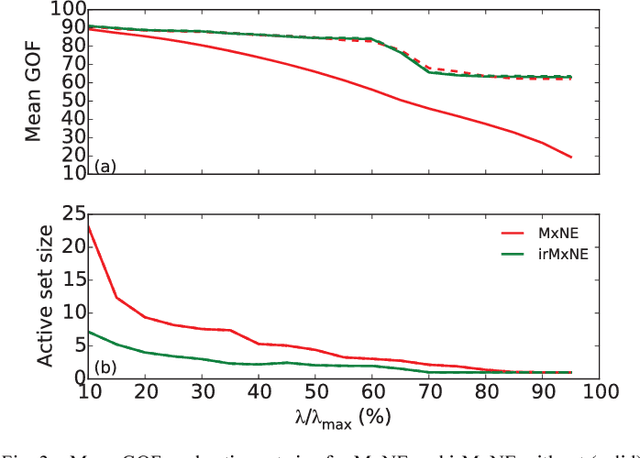
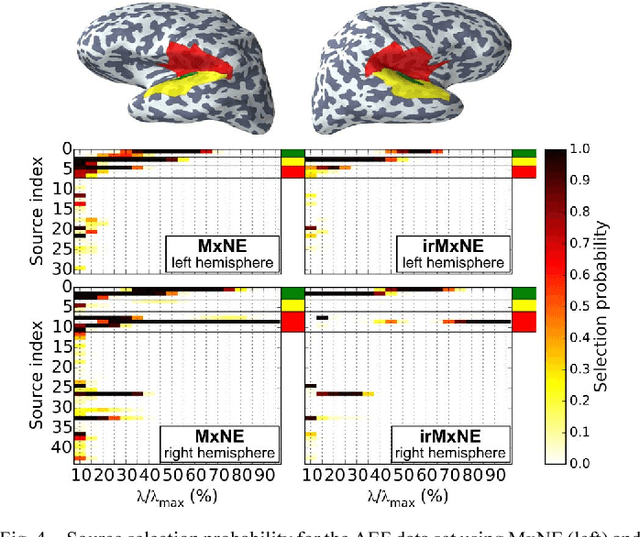
Abstract:Source imaging based on magnetoencephalography (MEG) and electroencephalography (EEG) allows for the non-invasive analysis of brain activity with high temporal and good spatial resolution. As the bioelectromagnetic inverse problem is ill-posed, constraints are required. For the analysis of evoked brain activity, spatial sparsity of the neuronal activation is a common assumption. It is often taken into account using convex constraints based on the l1-norm. The resulting source estimates are however biased in amplitude and often suboptimal in terms of source selection due to high correlations in the forward model. In this work, we demonstrate that an inverse solver based on a block-separable penalty with a Frobenius norm per block and a l0.5-quasinorm over blocks addresses both of these issues. For solving the resulting non-convex optimization problem, we propose the iterative reweighted Mixed Norm Estimate (irMxNE), an optimization scheme based on iterative reweighted convex surrogate optimization problems, which are solved efficiently using a block coordinate descent scheme and an active set strategy. We compare the proposed sparse imaging method to the dSPM and the RAP-MUSIC approach based on two MEG data sets. We provide empirical evidence based on simulations and analysis of MEG data that the proposed method improves on the standard Mixed Norm Estimate (MxNE) in terms of amplitude bias, support recovery, and stability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge