Jason Adams
Comparing the quality of neural network uncertainty estimates for classification problems
Aug 11, 2023Abstract:Traditional deep learning (DL) models are powerful classifiers, but many approaches do not provide uncertainties for their estimates. Uncertainty quantification (UQ) methods for DL models have received increased attention in the literature due to their usefulness in decision making, particularly for high-consequence decisions. However, there has been little research done on how to evaluate the quality of such methods. We use statistical methods of frequentist interval coverage and interval width to evaluate the quality of credible intervals, and expected calibration error to evaluate classification predicted confidence. These metrics are evaluated on Bayesian neural networks (BNN) fit using Markov Chain Monte Carlo (MCMC) and variational inference (VI), bootstrapped neural networks (NN), Deep Ensembles (DE), and Monte Carlo (MC) dropout. We apply these different UQ for DL methods to a hyperspectral image target detection problem and show the inconsistency of the different methods' results and the necessity of a UQ quality metric. To reconcile these differences and choose a UQ method that appropriately quantifies the uncertainty, we create a simulated data set with fully parameterized probability distribution for a two-class classification problem. The gold standard MCMC performs the best overall, and the bootstrapped NN is a close second, requiring the same computational expense as DE. Through this comparison, we demonstrate that, for a given data set, different models can produce uncertainty estimates of markedly different quality. This in turn points to a great need for principled assessment methods of UQ quality in DL applications.
Target Detection on Hyperspectral Images Using MCMC and VI Trained Bayesian Neural Networks
Aug 11, 2023Abstract:Neural networks (NN) have become almost ubiquitous with image classification, but in their standard form produce point estimates, with no measure of confidence. Bayesian neural networks (BNN) provide uncertainty quantification (UQ) for NN predictions and estimates through the posterior distribution. As NN are applied in more high-consequence applications, UQ is becoming a requirement. BNN provide a solution to this problem by not only giving accurate predictions and estimates, but also an interval that includes reasonable values within a desired probability. Despite their positive attributes, BNN are notoriously difficult and time consuming to train. Traditional Bayesian methods use Markov Chain Monte Carlo (MCMC), but this is often brushed aside as being too slow. The most common method is variational inference (VI) due to its fast computation, but there are multiple concerns with its efficacy. We apply and compare MCMC- and VI-trained BNN in the context of target detection in hyperspectral imagery (HSI), where materials of interest can be identified by their unique spectral signature. This is a challenging field, due to the numerous permuting effects practical collection of HSI has on measured spectra. Both models are trained using out-of-the-box tools on a high fidelity HSI target detection scene. Both MCMC- and VI-trained BNN perform well overall at target detection on a simulated HSI scene. This paper provides an example of how to utilize the benefits of UQ, but also to increase awareness that different training methods can give different results for the same model. If sufficient computational resources are available, the best approach rather than the fastest or most efficient should be used, especially for high consequence problems.
Deep Learning-Based Detection of the Acute Respiratory Distress Syndrome: What Are the Models Learning?
Sep 25, 2021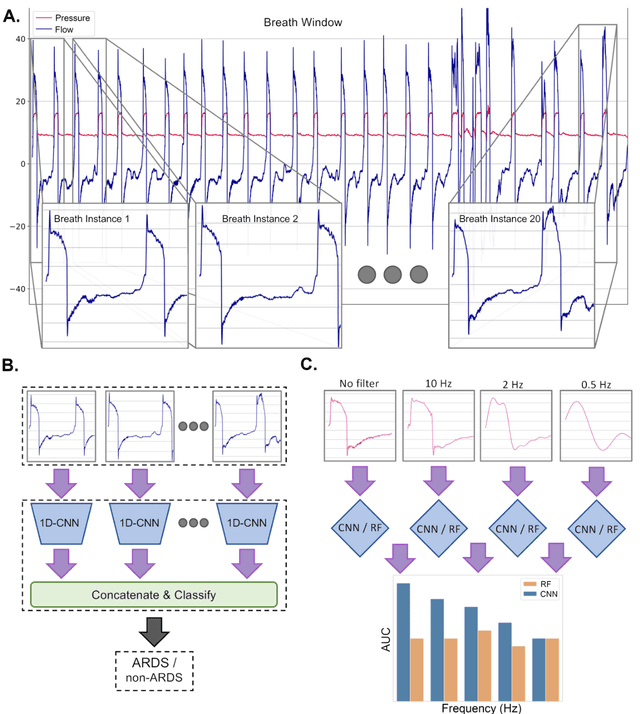
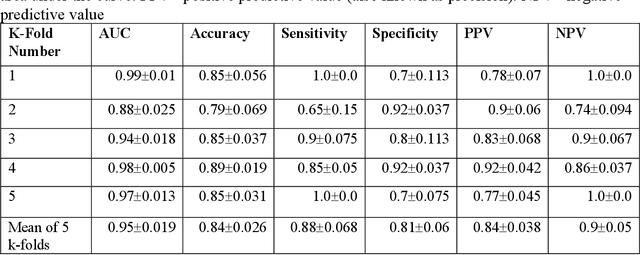
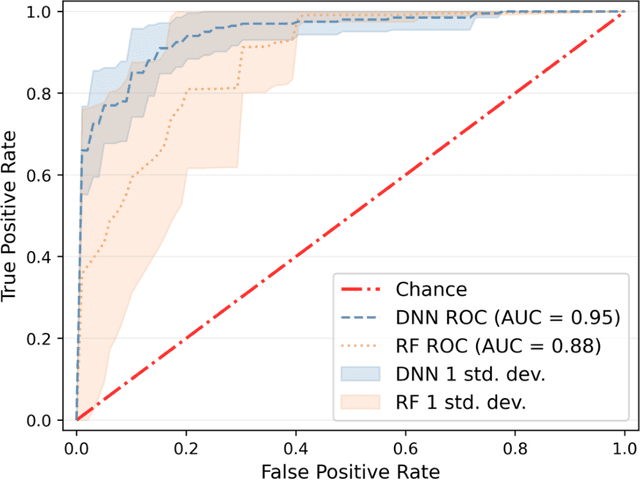
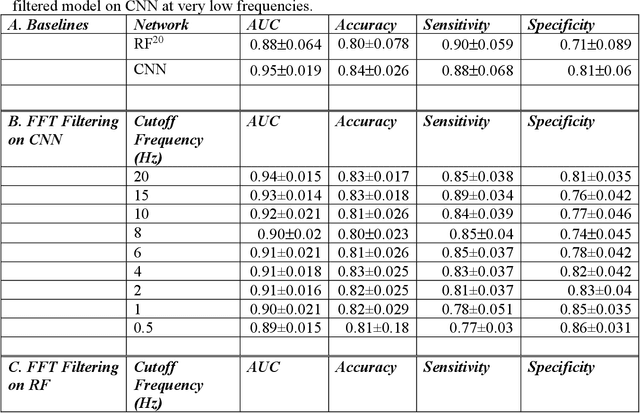
Abstract:The acute respiratory distress syndrome (ARDS) is a severe form of hypoxemic respiratory failure with in-hospital mortality of 35-46%. High mortality is thought to be related in part to challenges in making a prompt diagnosis, which may in turn delay implementation of evidence-based therapies. A deep neural network (DNN) algorithm utilizing unbiased ventilator waveform data (VWD) may help to improve screening for ARDS. We first show that a convolutional neural network-based ARDS detection model can outperform prior work with random forest models in AUC (0.95+/-0.019 vs. 0.88+/-0.064), accuracy (0.84+/-0.026 vs 0.80+/-0.078), and specificity (0.81+/-0.06 vs 0.71+/-0.089). Frequency ablation studies imply that our model can learn features from low frequency domains typically used for expert feature engineering, and high-frequency information that may be difficult to manually featurize. Further experiments suggest that subtle, high-frequency components of physiologic signals may explain the superior performance of DL models over traditional ML when using physiologic waveform data. Our observations may enable improved interpretability of DL-based physiologic models and may improve the understanding of how high-frequency information in physiologic data impacts the performance our DL model.
Clinical Validation of Single-Chamber Model-Based Algorithms Used to Estimate Respiratory Compliance
Sep 19, 2021



Abstract:Non-invasive estimation of respiratory physiology using computational algorithms promises to be a valuable technique for future clinicians to detect detrimental changes in patient pathophysiology. However, few clinical algorithms used to non-invasively analyze lung physiology have undergone rigorous validation in a clinical setting, and are often validated either using mechanical devices, or with small clinical validation datasets using 2-8 patients. This work aims to improve this situation by first, establishing an open, and clinically validated dataset comprising data from both mechanical lungs and nearly 40,000 breaths from 18 intubated patients. Next, we use this data to evaluate 15 different algorithms that use the "single chamber" model of estimating respiratory compliance. We evaluate these algorithms under varying clinical scenarios patients typically experience during hospitalization. In particular, we explore algorithm performance under four different types of patient ventilator asynchrony. We also analyze algorithms under varying ventilation modes to benchmark algorithm performance and to determine if ventilation mode has any impact on the algorithm. Our approach yields several advances by 1) showing which specific algorithms work best clinically under varying mode and asynchrony scenarios, 2) developing a simple mathematical method to reduce variance in algorithmic results, and 3) presenting additional insights about single-chamber model algorithms. We hope that our paper, approach, dataset, and software framework can thus be used by future researchers to improve their work and allow future integration of "single chamber" algorithms into clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge