Jan-Michaél Hirsch
Let it shine: Autofluorescence of Papanicolaou-stain improves AI-based cytological oral cancer detection
Jul 02, 2024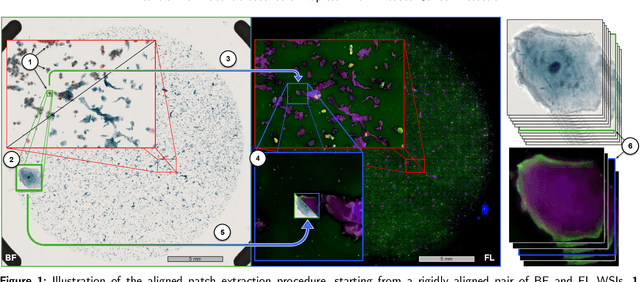
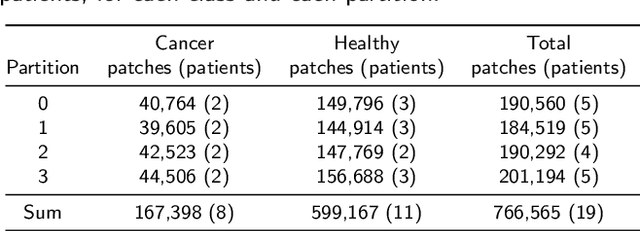
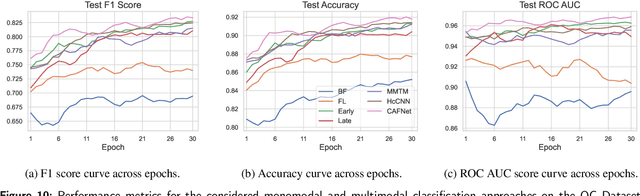
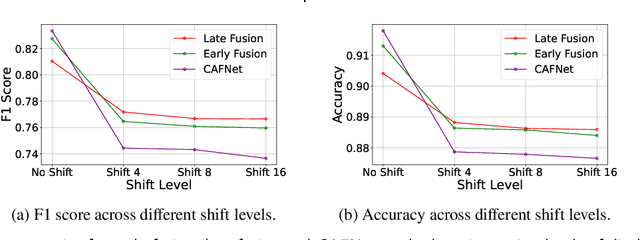
Abstract:Oral cancer is a global health challenge. It is treatable if detected early, but it is often fatal in late stages. There is a shift from the invasive and time-consuming tissue sampling and histological examination, toward non-invasive brush biopsies and cytological examination. Reliable computer-assisted methods are essential for cost-effective and accurate cytological analysis, but the lack of detailed cell-level annotations impairs model effectiveness. This study aims to improve AI-based oral cancer detection using multimodal imaging and deep fusion. We combine brightfield and fluorescence whole slide microscopy imaging to analyze Papanicolaou-stained liquid-based cytology slides of brush biopsies collected from both healthy and cancer patients. Due to limited cytological annotations, we utilize a weakly supervised deep learning approach using only patient-level labels. We evaluate various multimodal fusion strategies, including early, late, and three recent intermediate fusion methods. Our results show: (i) fluorescence imaging of Papanicolaou-stained samples provides substantial diagnostic information; (ii) multimodal fusion enhances classification and cancer detection accuracy over single-modality methods. Intermediate fusion is the leading method among the studied approaches. Specifically, the Co-Attention Fusion Network (CAFNet) model excels with an F1 score of 83.34% and accuracy of 91.79%, surpassing human performance on the task. Additional tests highlight the need for precise image registration to optimize multimodal analysis benefits. This study advances cytopathology by combining deep learning and multimodal imaging to enhance early, non-invasive detection of oral cancer, improving diagnostic accuracy and streamlining clinical workflows. The developed pipeline is also applicable in other cytological settings. Our codes and dataset are available online for further research.
A Deep Learning based Pipeline for Efficient Oral Cancer Screening on Whole Slide Images
Oct 23, 2019



Abstract:Oral cancer incidence is rapidly increasing worldwide. The most important determinant factor in cancer survival is early diagnosis. To facilitate large scale screening, we propose a fully automated end-to-end pipeline for oral cancer screening on whole slide cytology images. The pipeline consists of regression based nucleus detection, followed by per cell focus selection, and CNN based classification. We demonstrate that the pipeline provides fast and efficient cancer classification of whole slide cytology images, improving over previous results. The complete source code is made available as open source (https://github.com/MIDA-group/OralScreen).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge